Difference between revisions of "2016 Winter Project Week/Projects/ImageRestoration"
| (4 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
Image:PW-MIT2016.png|[[2016_Winter_Project_Week#Projects|Projects List]] | Image:PW-MIT2016.png|[[2016_Winter_Project_Week#Projects|Projects List]] | ||
Image:WMH_T1.png|Clinical Stroke Image | Image:WMH_T1.png|Clinical Stroke Image | ||
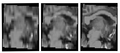
| + | Image:gmm_interp.png|Running result: Original (linear-interp) data | our gmm-based interp | "true" high res data | ||
| + | Image:gmmResult2.png|More results show our ability to improve images, but only on small areas. | ||
</gallery> | </gallery> | ||
| Line 17: | Line 19: | ||
To improve results for large datasets of clinical-quality data, we are investigating restoration methods without training datasets. Here, we are using a patch-based Gaussian Mixture Model approach with MRF priors and utilizing only the current dataset, without an external training dataset. | To improve results for large datasets of clinical-quality data, we are investigating restoration methods without training datasets. Here, we are using a patch-based Gaussian Mixture Model approach with MRF priors and utilizing only the current dataset, without an external training dataset. | ||
| + | |||
| + | <strong>Essentially, we explore a model where for a given location in all volumes of a dataset, we model those image patches as drawn from a particular mixture model.</strong> | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 28: | Line 32: | ||
| | | | ||
<!-- Add a bulleted list of key points --> | <!-- Add a bulleted list of key points --> | ||
| − | * | + | * We've implemented a small version of the model without regularization and that only works on a small section of a volume. We'll try to deploy it on large-scale dataset on entire volumes. |
| | | | ||
<!-- Fill this out at the end of Project Week; describe what you did this week and what you plan to do next --> | <!-- Fill this out at the end of Project Week; describe what you did this week and what you plan to do next --> | ||
| − | * | + | * We've built infrastructure for deploying the training at each location in parallele |
| + | * We've used our lab's cluster to deploy training with various parameter at each of 18,000 locations | ||
| + | * We've coded reconstruction modules to rebuild each region, but still need to combine each of the reconstructions. | ||
|} | |} | ||
Latest revision as of 14:09, 8 January 2016
Home < 2016 Winter Project Week < Projects < ImageRestorationKey Investigators
- Adrian Dalca (MIT)
- Katie Bouman (MIT)
- Polina Golland (MIT)
Project Description
Most synthesis, in-painting or super-resolution methods require a training dataset which includes the desired-quality images. Unfortunately, in the clinical setting this is often now available.
Due to the low quality of clinical images (often with many artifacts, 7mm thick slices, etc), most standard algorithms, such as those for registration, segmentation, analysis, will fail.
To improve results for large datasets of clinical-quality data, we are investigating restoration methods without training datasets. Here, we are using a patch-based Gaussian Mixture Model approach with MRF priors and utilizing only the current dataset, without an external training dataset.
Essentially, we explore a model where for a given location in all volumes of a dataset, we model those image patches as drawn from a particular mixture model.
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
|