Difference between revisions of "2017 Winter Project Week/LiverResectionPlanning"
From NAMIC Wiki
Louise.oram (talk | contribs) |
Louise.oram (talk | contribs) |
||
| (8 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-Winter2017.png|link=2017_Winter_Project_Week#Projects|[[2017_Winter_Project_Week#Projects|Projects List]] | Image:PW-Winter2017.png|link=2017_Winter_Project_Week#Projects|[[2017_Winter_Project_Week#Projects|Projects List]] | ||
| + | Image:Screen_Shot_2017-01-09_at_1.23.51_PM.png | ||
| + | Image:Screen_Shot_2017-01-09_at_1.22.47_PM.png | ||
<!-- Use the "Upload file" link on the left and then add a line to this list like "File:MyAlgorithmScreenshot.png" --> | <!-- Use the "Upload file" link on the left and then add a line to this list like "File:MyAlgorithmScreenshot.png" --> | ||
| + | |||
</gallery> | </gallery> | ||
| Line 8: | Line 11: | ||
<!-- Add a bulleted list of investigators and their institutions here --> | <!-- Add a bulleted list of investigators and their institutions here --> | ||
* Louise Oram, The intervention centre - Oslo University Hospital | * Louise Oram, The intervention centre - Oslo University Hospital | ||
| + | * Andrey Fedorov, BWH | ||
| + | * Christian Herz, BWH | ||
| + | * Andras Lasso, Queens | ||
==Project Description== | ==Project Description== | ||
| Line 17: | Line 23: | ||
| | | | ||
<!-- Objective bullet points --> | <!-- Objective bullet points --> | ||
| − | * To create an extension that allows surgeons to create 3D models and plan liver resection surgeries | + | * To create an extension that allows surgeons to create 3D models and plan liver resection surgeries. |
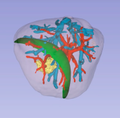
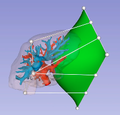
* This extension will contain modules for segmenting parts of the liver (parenchyma, tumours, portal and hepatic veins), as well as a module for creating a resection plan in the resulting 3D model. | * This extension will contain modules for segmenting parts of the liver (parenchyma, tumours, portal and hepatic veins), as well as a module for creating a resection plan in the resulting 3D model. | ||
| | | | ||
| Line 25: | Line 31: | ||
| | | | ||
<!-- Progress and Next steps bullet points (fill out at the end of project week) --> | <!-- Progress and Next steps bullet points (fill out at the end of project week) --> | ||
| − | * | + | * Using VTK field data to store / retrieve extra information for resection surfaces |
| − | * | + | * Structure in place for loading / saving resection surfaces (resectionSurfaceNodes inheriting from modelNode, reader, registering IO...) |
| + | Fruitful discussions: | ||
| + | * Learnt more about options for customizing Slicer | ||
| + | * Plan to use SegmentationNode to solve issues with needing to load many models | ||
| + | * Feedback on modules, should try to generalize the vessel segmentation to be usable on other images (not just contrast enhanced CT of the liver) | ||
|} | |} | ||
Latest revision as of 16:11, 13 January 2017
Home < 2017 Winter Project Week < LiverResectionPlanningKey Investigators
- Louise Oram, The intervention centre - Oslo University Hospital
- Andrey Fedorov, BWH
- Christian Herz, BWH
- Andras Lasso, Queens
Project Description
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
Fruitful discussions:
|