Difference between revisions of "2017 Winter Project Week/WhiteMatterAnalysis"
From NAMIC Wiki
| Line 4: | Line 4: | ||
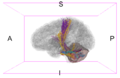
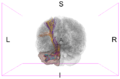
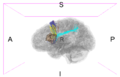
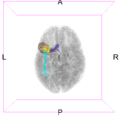
Image:Left CB.png|Left-view Cerebellum | Image:Left CB.png|Left-view Cerebellum | ||
Image:Post CB.png|Posterior-view Cerebellum | Image:Post CB.png|Posterior-view Cerebellum | ||
| − | Image:Left ParsOp.png|Left-view Pars | + | Image:Left ParsOp.png|Left-view Pars Opercularis |
| − | Image:Sup ParsOp.png|Superior-view Pars | + | Image:Sup ParsOp.png|Superior-view Pars Opercularis |
<!-- Use the "Upload file" link on the left and then add a line to this list like "File:MyAlgorithmScreenshot.png" --> | <!-- Use the "Upload file" link on the left and then add a line to this list like "File:MyAlgorithmScreenshot.png" --> | ||
Latest revision as of 15:53, 13 January 2017
Home < 2017 Winter Project Week < WhiteMatterAnalysisKey Investigators
- Fan Zhang, BWH
- Shun Gong, BWH
- Isaiah Norton, BWH
- Ye Wu, BWH
- Lauren J. O'Donnell, BWH
Project Description
This project is part of the whitematteranalysis software that provides an automated whole brain tractgoraphy registration and segmentation framework. The aim of this project is to integrate a new Slicer module FiberEndpointAnalysis into the whitematteranalysis pipeline.
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
Calculate the overlap of fiber tract (endpoint regions) with anatomical parcellation (e.g. freesurfer) or functional region (e.g. fMRI). |
|
|
Background and References
- SlicerDMRI: http://slicerdmri.github.io
- WhiteMatterAnalysis: https://github.com/SlicerDMRI/whitematteranalysis