Difference between revisions of "AHM2010:Mind"
Hjbockholt (talk | contribs) |
Hjbockholt (talk | contribs) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 14: | Line 14: | ||
==MRN Roadmap Project== | ==MRN Roadmap Project== | ||
| + | |||
| + | == Overview == | ||
| + | * What problem does the pipeline solve, and who is the targeted user? | ||
| + | **The pipeline attempts to solve the problem of segmenting white matter lesions in Neuropsychiatric Systemic Lupus Erythematosus(NPSLE). The automated capability is aimed at clinical researchers using Slicer3 software. The utility of having accurate lesion labels permits summary of perfusion within lesions, correlation of lesion load by location with neuropsychiatric symptoms, and summary of structural image intensities or DTI scalars within lesion boundaries. | ||
| + | |||
| + | * How does the pipeline compare to state of the art? | ||
| + | **Brain lesion classification in NPSLE is currently accomplished manually or by various algorithms developed for other disorders, such as multiple sclerosis (MS).There are many approaches that attempt to solve lesion classification in MS. Some of these approaches are automated or semi-automated; however, all automatic approaches suffer a lack of ground truth. It is difficult for human manual raters to agree on fuzzy boundaries across different image contrasts (e.g., T1, T2, FLAIR). | ||
| + | |||
| + | A good examples of the challenges that remains in terms of lesion segmentation were represented at the 2008 MICCAI Conference [http://www.ia.unc.edu/MSseg/ MS Lesion Segmentation Challenge(organized by Warfield and Styner)] in the form of a grand challenge in segmentation. Our participation in this lesion segmentation contest, in which participants were provided training data-sets with two manual labellings, allowed us to experiment with several approaches to lesion segmentation, e.g., EM-Segment (S. Wells), k-means/bayesian (V. Magnotta), and outlier detection (M. Prastawa). Drawing from this experience and a thorough review of the current literature, we have optimized a novel combination of methods that appears to perform well for lesion mapping in NPSLE. Additionally, this approach may generalize to other neurological disorders, such as MS or vascular dementia. | ||
| + | |||
| + | |||
{| | {| | ||
| Line 24: | Line 35: | ||
|} | |} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ||
==Detailed Information about the Pipeline== | ==Detailed Information about the Pipeline== | ||
| Line 74: | Line 80: | ||
{| | {| | ||
|[[Image:CompareViewFlairLesionDiffWholeBrain.png|thumb|333px|CompareViewFlairLesionDiffWholeBrain]] | |[[Image:CompareViewFlairLesionDiffWholeBrain.png|thumb|333px|CompareViewFlairLesionDiffWholeBrain]] | ||
| − | |||
|[[Image:CompareViewFlairLesionDiff.png|thumb|333px|CompareViewFlairLesionDiff]] | |[[Image:CompareViewFlairLesionDiff.png|thumb|333px|CompareViewFlairLesionDiff]] | ||
|} | |} | ||
Latest revision as of 16:10, 7 January 2010
Home < AHM2010:MindTeam
- DBP: H Jeremy Bockholt (PI), Charles Gasparovic(co-PI), Mark Scully(Engineer), The Mind Research Network
- Core 2: Steve Pieper, Isomics
- Contact: H. Jeremy Bockholt, jbockholt@mrn.org
Lupus
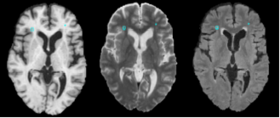
Systemic lupus erythematosus (SLE) is an autoimmune disease affecting multiple tissues, including the brain the facial rash of some people with lupus looked like the bite or scratch of a wolf ("lupus" is Latin for wolf and "erythematosus" is Latin for red). patients may feel weak and fatigued, have muscle aches, loss of appetite, swollen glands, and hair loss, sometimes have abdominal pain, nausea, diarrhea, and vomiting. Estimates of SLE prevalence range from 14.6-372 per 105 About 1.5 million americans, 90% diagnosed are female Neuropsychiatric SLE (NPSLE), a term that subsumes the neurologic and psychiatric complications of SLE, occurs in up to 95% of SLE patients While MRI often reveals distinct white matter abnormalities in active NPSLE, the pathologic processes underlying these lesions, whether purely autoimmune or vascular (e.g., hemostasis), are unknown
MRN Roadmap Project
Overview
- What problem does the pipeline solve, and who is the targeted user?
- The pipeline attempts to solve the problem of segmenting white matter lesions in Neuropsychiatric Systemic Lupus Erythematosus(NPSLE). The automated capability is aimed at clinical researchers using Slicer3 software. The utility of having accurate lesion labels permits summary of perfusion within lesions, correlation of lesion load by location with neuropsychiatric symptoms, and summary of structural image intensities or DTI scalars within lesion boundaries.
- How does the pipeline compare to state of the art?
- Brain lesion classification in NPSLE is currently accomplished manually or by various algorithms developed for other disorders, such as multiple sclerosis (MS).There are many approaches that attempt to solve lesion classification in MS. Some of these approaches are automated or semi-automated; however, all automatic approaches suffer a lack of ground truth. It is difficult for human manual raters to agree on fuzzy boundaries across different image contrasts (e.g., T1, T2, FLAIR).
A good examples of the challenges that remains in terms of lesion segmentation were represented at the 2008 MICCAI Conference MS Lesion Segmentation Challenge(organized by Warfield and Styner) in the form of a grand challenge in segmentation. Our participation in this lesion segmentation contest, in which participants were provided training data-sets with two manual labellings, allowed us to experiment with several approaches to lesion segmentation, e.g., EM-Segment (S. Wells), k-means/bayesian (V. Magnotta), and outlier detection (M. Prastawa). Drawing from this experience and a thorough review of the current literature, we have optimized a novel combination of methods that appears to perform well for lesion mapping in NPSLE. Additionally, this approach may generalize to other neurological disorders, such as MS or vascular dementia.
Detailed Information about the Pipeline
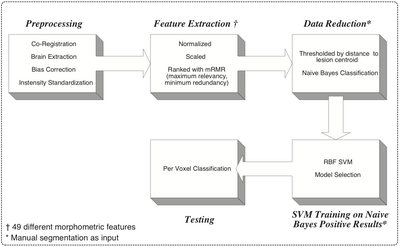
- Method
- An Automated Method For Segmenting White Matter Lesions in Lupus Through Multi-level Morphometric Feature Classification
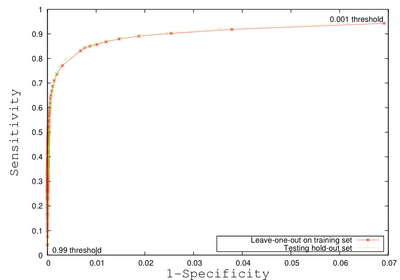
- The method makes use of local morphometric features based on multiple MR sequences, including T1-weighted, T2-weighted, and Fluid Attenuated Inversion Recovery. After preprocessing, including co-registration, brain extraction, bias correction, and intensity standardization, 49 features are calculated for each brain voxel based on local morphometry. At each level of segmenta- tion a supervised classifier takes advantage of a different subset of the features to conservatively segment lesion voxels, passing on more difficult voxels to the next classifier. This multi-level approach allows for a fast lesion classification method with tunable trade-offs between sensitivity and specificity producing accuracy comparable to a human rater.
- An Automated Method For Segmenting White Matter Lesions in Lupus Through Multi-level Morphometric Feature Classification
Software & documentation
- We have a very active project for this pipeline on the NITRC resource 3DSlicerLupusLesionModule
Outreach
- An end-to-end tutorial and module are provided on the NA-MIC website. Training courses were held at the Annual meeting for Human Brain Mapping Organization 2008, the Annual Meeting for Society for Neuroscience 2008, as well as to a group of 25+ investigators at the MIND Research Network (2007) who are working in translational neuroscience. A Manuscript summarizing improved clinical results resulting from automated lesion analyses has been prepared and submitted at the time of writing this report.
- Publication Links to the PubDB.
- H. J. Bockholt, V. A. Magnotta, M. Scully, C. Gasparovic, B. Davis, K. Pohl, R. Whitaker, S. Pieper, C. Roldan, R. Jung, R. Hayek, W. Sibbitt, J. Sharrar, P. Pellegrino, R. Kikinis. A novel automated method for classification of white matter lesions in systemic lupus erythematosus. Presented at the 38th annual meeting of the Society for Neuroscience, Washingto, DC, 15 – 19 November 2008
- Scully M., Magnotta V., Gasparovic C., Pelligrimo P., Feis D., Bockholt H.J. 3D Segmentation In The Clinic: A Grand Challenge II at MICCAI 2008 - MS Lesion Segmentation. IJ - 2008 MICCAI Workshop - MS Lesion Segmentation. Available http://grand-challenge2008.bigr.nl/proceedings/pdfs/msls08/282_Scully.pdf
- H Jeremy Bockholt, Josef Ling, Mark Scully, Adam Scott, Susan Lane, Vincent Magnotta, Tonya White, Kelvin Lim, Randy Gollub, Vince Calhoun. Real-time Web-scale Image Annotation for Semantic-based Retrieval of Neuropsychiatric Research Images. Presented at the 14th Annual Meeting of the Organization for Human Brain Mapping, Melbourne, Australia, 15 – 19 June, 2008.
- H Jeremy Bockholt, Sumner Williams, Mark Scully, Vincent Magnotta, Randy Gollub, John Lauriello, Kelvin Lim, Tonya White, Rex Jung, Charles Schulz, Nancy Andreasen, Vince Calhoun. The MIND Clinical Imaging Consortium as an application for novel comprehensive quality assurance procedures in a multi-site heterogeneous clinical research study. Presented at the 14th Annual Meeting of the Organization for Human Brain Mapping, Melbourne, Australia, 15 – 19 June, 2008.
- M Scully, B H Anderson, C Gasparovic, V A Magnotta, S Pieper, R Kikinis, P Pellegrino, T Lane, H J Bockholt. A Synergistic Combination of Supervised Machine Learning Methods for Analysis of White Matter Lesions in Neuropsychiatric Systemic Lupus Erythematosus. Presented at the 15th Annual Meeting of the Organization for Human Brain Mapping, San Francisco June, 2009.
- Bockholt HJ, Scully M, Courtney W, Rachakonda S, Scott A, Caprihan A, Fries J, Kalyanam R, Segall J, de la Garza R, Lane S and Calhoun VD (2009) Mining the mind research network: a novel framework for exploring large scale, heterogeneous translational neuroscience research data sources. Front. Neuroinform. 3:36. doi:10.3389/neuro.11.036.2009
- Scully, M., Anderson, B., Lane, T., Gasparovic, C., Magnotta, V., Sibbitt, W., Roldan,C. , Kikinis, R., Bockholt, H. J. An Automated Method For Segmenting White Matter Lesions in Lupus Through Multi-level Morphometric Feature Classification (submitted to Frontiers in Neuroscience 11/2009).
- Scully, M., Anderson, B., Lane, T., Gasparovic, C., Magnotta, V., Sibbitt, W., Roldan,C. , Kikinis, R., Bockholt, H. J. An Automated Method For Longitudinal Analysis of White Matter Lesions in Lupus (in preparation for submission).
- Bockholt, H.J., Gasparovic, C., Scully, M., Magnotta, V., Sibbitt, W., Kikinis, R., Roldan,C. A novel white matter lesion analysis for improved clinical application in lupus. (in preparation for submission).
In Progress since last AHM
- Longitudinal Analyses
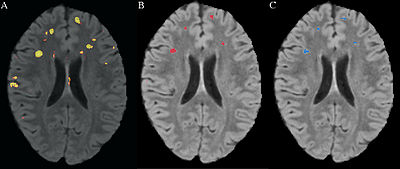
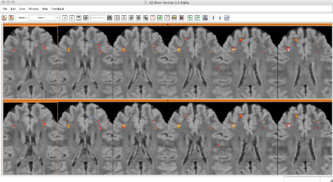
- Almost completed full pipeline for longitudinal analysis which detects changes in lesions allowing for more clinical applications. The following 3 figures demonstrate some of the longitudinal functionality comparing a baseline FLAIR with a followup FLAIR. The label map shows the change in lesion voxels where gained lesion is red, recovered tissue is blue, and lesion that remained lesion is yellow.
- DWI/DTI Analyses
- Population differences in Scalar values
- Tract based statistics
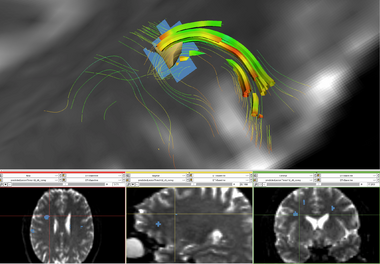
- Identifying structural connectivity network
- Approximating Disrupted Tractography Fibers
- Similar in purpose to current methods being applied to tumors, the objective is to estimate the tracts the would exist were lesion not present.
- Morphometry
- Examine cortical thicknesses especially in regions where lesions have interrupted connecting fibers.
- Investigate relationship between diseased tissue and fractal dimension of the white matter boundary.
- Spectroscopic Imaging
- Functional rest task
- Functional Network
- ASL
- Perfusion
- Relationship between cerebral blood volume, cerebral blood flow, and NPSLE
- Perfusion
- Multiscale Analyses
- Incorporate all previous results into a comprehensive statistical analysis to fully characterize NPSLE.
Future Collaboration
Two grants are under preparation to continue this collaboration. The first grant in progress with a planned submission of Feb 5, 2010 will use the Collaborations with NCBC R01 mechanism. The grant will be titled, “Enhancing Brain Lesion Segmentation in Neurological Disorders.” The aim of this project will be to extend the tools developed in the lupus DBP to other vascular disorders, multiple sclerosis, vascular dementia and other disorders with brain lesions. The grant will be a natural extension of work in the lupus DBP and fits in well with the mission of the NA-MIC by extending tools, analyses and approaches across multiple diseases and challenges.
The second grant will utilize the GPL-style license found within the NA-MIC to explore the commercialization potential of the lesion analysis toolkit. A grant entitled “Novel White Matter Lesion Application” has a planned submission of April 5, 2010 and will use the Neurotechnology Research, Development, and Enhancement SBIR mechanism. This grant will be submitted by PI Bockholt who has recently founded and formed Advanced Biomedical Informatics Group, LLC, an independent for-profit company based in Iowa City, IA. This project will also extend the lupus DBP work through further development of turnkey solution that can be placed directly in the hands of clinicians for evaluating white matter lesions in their lupus patients
References
- Adaboost and Support Vector Machines for White Matter Lesion Segmentation in MR Images. Quddus A, Fieguth P, Basir O. PAMI Lab, Department of Electrical and Computer Engineering, University of Waterloo, Waterloo, ON, Canada. Conf Proc IEEE Eng Med Biol Soc. 2005;1:463-6. http://www.ncbi.nlm.nih.gov/pubmed/17282216
- Architecture for an Artificial Immune System Steven A. Hofmeyr, Stephanie Forrest Evolutionary Computation Winter 2000, Vol. 8, No. 4, Pages 443-473 http://www.mitpressjournals.org/doi/abs/10.1162/106365600568257
- Automated segmentation of white matter lesions in 3D brain MR images, using multivariate pattern classification Zhiqiang Lao; Dinggang Shen; Jawad, A.; Karacali, B.; Dengfeng Liu; Melhem, E.R.; Bryan, R.N.; Davatzikos, C. Biomedical Imaging: Nano to Macro, 2006. 3rd IEEE International Symposium on Volume , Issue , 6-9 April 2006 Page(s): 307 - 310 http://ieeexplore.ieee.org/Xplore/login.jsp?url=/iel5/10818/34114/01624914.pdf?temp=x
- Automatic Segmentation of MS Lesions Using a Contextual Model for the MICCAI Grand Challenge Jonathan H. Morra, Zhuowen Tu, Arthur W. Toga, Paul M. Thompson IJ - 2008 MICCAI Workshop - MS Lesion Segmentation http://www.midasjournal.org/browse/publication/280