Difference between revisions of "Algorithm:UNC"
| Line 1: | Line 1: | ||
| − | + | Back to [[Algorithm:Main|NA-MIC Algorithms]] | |
__NOTOC__ | __NOTOC__ | ||
= Overview of UNC Algorithms (PI: Martin Styner) = | = Overview of UNC Algorithms (PI: Martin Styner) = | ||
| Line 19: | Line 19: | ||
<font color="red">'''New: '''</font> | <font color="red">'''New: '''</font> | ||
| − | * Co-Organization of MICCAI | + | * Co-Organization of MICCAI 2008 workshop on [http://grand-challenge2008.bigr.nl "3D Segmentation in the Clinic II- A Grand Challenge"] with a competition on Multiple Sclerosis lesion segmentation |
| − | * Continuation of workshop competition on [http:// | + | * Continuation of workshop competition on the [http://www.ia.unc.edu/MSseg/ MS segmentation online comparison] |
| − | |||
|- | |- | ||
Revision as of 20:20, 29 October 2008
Home < Algorithm:UNCBack to NA-MIC Algorithms
Overview of UNC Algorithms (PI: Martin Styner)
At UNC, we are interested in a range of algorithms and solutions for the surface based analysis of brain structures and the cortex. We pioneered the use of spherical harmonics based shape analysis for comparing brain structures across objects. We are now working on incorporating various data sources on the entire cortical surface for improving the correspondence computation. Furthermore, validation and evaluation of methods is highly relevant within our core.
UNC Projects

|
Evaluation and Comparison of Medical Image Analysis MethodsIn this project, we want to focus on the evaluation of medical image analysis methods for specific clinical applications in respect to development of evaluation methodology and the organization of venues promoting such comparison and validation studies. New:
|
|
Cortical Correspondence using Particle SystemIn this project, we want to compute cortical correspondence on populations, using various features such as cortical structure, DTI connectivity, vascular structure, and functional data (fMRI). This presents a challenge because of the highly convoluted surface of the cortex, as well as because of the different properties of the data features we want to incorporate together. More... New:
|

|
Shape Analysis Framework using SPHARM-PDMThe UNC shape analysis is based on an analysis framework of objects with spherical topology, described mainly by sampled spherical harmonics SPHARM-PDM. The input of the shape analysis framework is a set of binary segmentations of a single brain structure, such as the hippocampus or caudate. These segmentations are converted into a shape description (SPHARM) with correspondence and analyzed via Hotelling T^2 two sample metric. More... New:
|

|
Population Based CorrespondenceWe are developing methodology to automatically find dense point correspondences between a collection of polygonal genus 0 meshes. The advantage of this method is independence from indivisual templates, as well as enhanced modeling properties. The method is based on minimizing a cost function that describes the goodness of correspondence. Apart from a cost function derived from the description length of the model, we also employ a cost function working with arbitrary local features. We extended the original methods to use surface curvature measurements, which are independent to differences of object aligment. More... New:
|
|
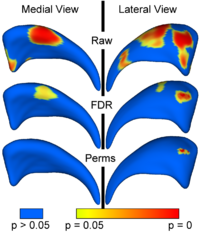
Local Statistical Analysis via Permutation TestsWe have further developed a set of statistical testing methods that allow the analysis of local shape differences using the Hotelling T 2 two sample metric. Permutatioin tests are employed for the computation of statistical p-values, both raw and corrected for multiple comparisons. Resulting significance maps are easily visualized. Additional visualization of the group tests are provided via mean difference magnitude and vector maps, as well as maps of the group covariance information. Ongoing research focuses on incorporating covariates such as clinical scores into the testing scheme. More... New:
|