Difference between revisions of "Algorithm:UNC"
| Line 84: | Line 84: | ||
== [[Projects:ShapeAnalysisFrameworkUsingSPHARMPDM|UNC-Utah Shape Analysis Framework]] == | == [[Projects:ShapeAnalysisFrameworkUsingSPHARMPDM|UNC-Utah Shape Analysis Framework]] == | ||
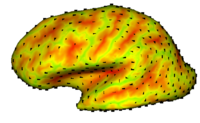
| − | The UNC shape analysis is based on an analysis framework of objects with spherical topology, described mainly by sampled spherical harmonics SPHARM-PDM. The input of the shape analysis framework is a set of binary segmentations of a single brain structure, such as the hippocampus or caudate. These segmentations are converted into a shape description (SPHARM-PDM) with correspondence and tested via statistical point-wise analysis. [[Projects:ShapeAnalysisFrameworkUsingSPHARMPDM|More...]] | + | The UNC shape analysis is based on an analysis framework of objects with spherical topology, described mainly by sampled spherical harmonics SPHARM-PDM. The input of the shape analysis framework is a set of binary segmentations of a single brain structure, such as the hippocampus or caudate. These segmentations are converted into a shape description (SPHARM-PDM) with correspondence and tested via statistical point-wise analysis. Additionally, the SPHARM correspondences can be improved with Entropy-based Shape Analysis system, by using an integration module recently added to the pipeline. [[Projects:ShapeAnalysisFrameworkUsingSPHARMPDM|More...]] |
| − | * Shape Analysis Toolkit disseminated on [http://www.nitrc.org/projects/spharm-pdm NITRC SPHARM PDM page]. All tools are Slicer compatible. | + | * SPHARM-Particle Shape Analysis Toolkit disseminated on [http://www.nitrc.org/projects/spharm-pdm NITRC SPHARM PDM page]. All tools are Slicer compatible. |
* Single Slicer 3 module for whole shape analysis pipeline | * Single Slicer 3 module for whole shape analysis pipeline | ||
Revision as of 15:36, 24 October 2011
Home < Algorithm:UNCBack to NA-MIC Algorithms
Overview of UNC Algorithms (PI: Martin Styner)
At UNC, we are interested in a range of algorithms and solutions for the surface based analysis of brain structures and the cortex. We pioneered the use of spherical harmonics based shape analysis for comparing brain structures across objects. We has also worked on incorporating various data sources for correspondence computation on surfaces of different complexity (ranging from simple brain structures to the highly folded cortical surface). A current topic includes the use of diffusion tensor imaging for connectivity analysis in pathological settings. Finally, investigating quality control, validation and evaluation methodology is another important topic of our NA-MIC research.
UNC Projects
|
Longitudinal Atlas BuildingAs part of the longitudinal intra- and interpatient analysis theme within NA-MIC, we are working on a deformable, longitudinal DTI atlas method. Our longitudinal framework explicitly accounts for temporal dependencies via iterative subject-specific statistical growth modeling, and cross-sectional atlas-building. To effectively account for measurements sparse in time, a continuous-discrete statistical growth model is proposed incorporating also patient co-variatesMore...
|
|
Diffusion Weighted Atlas Construction via model-based transformation and averaging of signalThis project investigated a method for model-based averaging of sets of diffusion weighted magnetic resonance images (DW-MRI) under space transformations (resulting for example from registration methods). A robust weighted least squares method is developed. Synthetic validation experiments show the improvement of the proposed estimation method in comparison to standard least squares estimation. The developed method is applied to construct an atlas of {\it diffusion weighted images} for a set of macaques, allowing for a more flexible representation of average diffusion information compared to standard diffusion tensor atlases. More...
New: M. Niethammer, Y. Shi, S. Benzaid, M. Sanchez, and M. Styner. Robust model-based transformation and averaging of diffusion weighted images applied to diffusion weighted atlas construction. MICCAI, Workshop on Computational Diffusion MRI, 2010. |

|
Diffusion Imaging based ConnectivityThis project focuses on connectivity measurements derived from diffusion imaging datasets in order to better understand cortical and subcortical white matter connectivity. Our research employs a novel, multi-directional graph propagation method that performs a fully deterministic, efficient and stable connectivity computation. The method handles crossing fibers and deals well with multiple seed regions. In addition to the analysis of these connectivity measures in describing brain pathology, they can also be used as scalar maps for use in DTI registration. More...
|
|
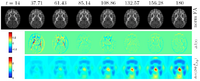
DWI and DTI Quality ControlDWI data suffers from inherent low SNR, overall long scanning time of multiple directional encoding with correspondingly large risk to encounter several kinds of artifacts. These artifacts can be too severe for a correct and stable estimation of the diffusion tensor. Thus, a quality control (QC) procedure is absolutely necessary for DTI studies. We are developing a framework for automatic DWI and DTI quality assessment and correction. We developed a tool called DTIPrep which pipelines the QC steps with designated protocol use and report generation. More... New: DTIPrep first full version on NITRC New: Zhexing Liu, Casey Goodlett, Guido Gerig, Martin Styner. Evaluation of DTI property maps as basis of DTI atlas building. Medical Imaging 2010: Image Processing (2010) vol. 7623 (1) pp. 762325 New: Zhexing Liu, Yi Wang, Guido Gerig, Sylvain Gouttard, Ran Tao, Thomas Fletcher, Martin Styner. Quality control of diffusion weighted images. Medical Imaging 2010: Advanced PACS-based Imaging Informatics and Therapeutic Applications (2010) vol. 7628 (1) pp. 76280J
|
|
Cortical Correspondence using Particle SystemIn this project, we want to compute cortical correspondence on populations, using various features such as cortical structure, DTI connectivity, vascular structure, and functional data (fMRI). This presents a challenge because of the highly convoluted surface of the cortex, as well as because of the different properties of the data features we want to incorporate together. This correspondence method has been included in our NAMIC cortical thickness framework GAMBIT More... New: Vachet, C., Hazlett, H., Niethammer, M., Oguz, I., Cates, J., Whitaker, R., Piven, J., Styner, M., “Group-wise automatic mesh-based analysis of cortical thickness“. Medical Imaging 2011: Image Processing (2011) vol. 7962 (1) pp. 796227 1 - 10 New: Lee J, Ehlers C, Crews F, Niethammer M, Budin F, Paniagua B, Sulik K, Johns J, Styner M, Oguz I. Automatic cortical thickness analysis on rodent brain. Medical Imaging 2011: Image Processing (2011) vol. 7962 (1) pp. 796248 |

|
UNC-Utah Shape Analysis FrameworkThe UNC shape analysis is based on an analysis framework of objects with spherical topology, described mainly by sampled spherical harmonics SPHARM-PDM. The input of the shape analysis framework is a set of binary segmentations of a single brain structure, such as the hippocampus or caudate. These segmentations are converted into a shape description (SPHARM-PDM) with correspondence and tested via statistical point-wise analysis. Additionally, the SPHARM correspondences can be improved with Entropy-based Shape Analysis system, by using an integration module recently added to the pipeline. More...
New: automatic generation of Slicer MRML scenes for result visualization New: Mark Walterfang, Jeffrey Chee Leong Looi, Martin Styner, Ruth H Walker, Adrian Danek, Marc Neithammer, Andrew Evans, Katya Kotschet, Guilherme R Rodrigues, Andrew Hughes, Dennis Velakoulis. Shape alterations in the striatum in chorea-acanthocytosis. Psychiatry research (2011) preprint online New: Beatriz Paniagua, Lucia Cevidanes, David Walker, Hongtu Zhu, Ruixin Guo, Martin Styner. Clinical application of SPHARM-PDM to quantify temporomandibular joint osteoarthritis. Computerized medical imaging and graphics : the official journal of the Computerized Medical Imaging Society (2011) preprint online. New: Beatriz Paniagua, Lucia Cevidanes, Hongtu Zhu, Martin Styner. Outcome quantification using SPHARM-PDM toolbox in orthognathic surgery. International journal of computer assisted radiology and surgery (2011) preprint online New: Jeffrey Chee Leong Looi, Mark Walterfang, Martin Styner, Leif Svensson, Olof Lindberg, Per Ostberg, Lisa Botes, Eva Orndahl, Phyllis Chua, Rajeev Kumar, Dennis Velakoulis, Lars-Olof Wahlund. Shape analysis of the neostriatum in frontotemporal lobar degeneration, Alzheimer's disease, and controls. Neuroimage (2010) vol. 51 (3) pp. 970-86
|

|
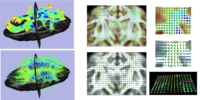
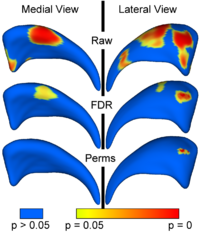
Local Statistical Analysis via Permutation TestsWe have further developed a set of statistical testing methods that allow the analysis of local shape differences via group differences tests as well interaction tests. Resulting significance maps (both raw and corrected for multiple comparisons) are easily visualized. Additional visualization of the group tests are provided via mean difference magnitude and vector maps, as well as maps of the group covariance information. Additional visualization of the interaction tests include Pearson and Spearman correlation maps. More... New: User-friendly GUI interface and statistical result visualization via automatically generated Slicer MRML scenes New: Available on NITRC either separately (ShapeAnalysisMANCOVA) or as part of the SPHARM-PDM shape analysis package
|

|
Evaluation and Comparison of Medical Image Analysis MethodsIn this project, we want to focus on the evaluation of medical image analysis methods for specific clinical applications in respect to development of evaluation methodology and the organization of venues promoting such comparison and validation studies. New: DTI fiber tractography challenge at MICCAI 2011 |
|
Population Based CorrespondenceWe are developing methodology to automatically find dense point correspondences between a collection of polygonal genus 0 meshes. The advantage of this method is independence from indivisual templates, as well as enhanced modeling properties. The method is based on minimizing a cost function that describes the goodness of correspondence. Apart from a cost function derived from the description length of the model, we also employ a cost function working with arbitrary local features. We extended the original methods to use surface curvature measurements, which are independent to differences of object aligment. More...
|