Difference between revisions of "Algorithm:UNC:DTI:Population Analysis"
(added feature images) |
|||
| Line 1: | Line 1: | ||
= Description = | = Description = | ||
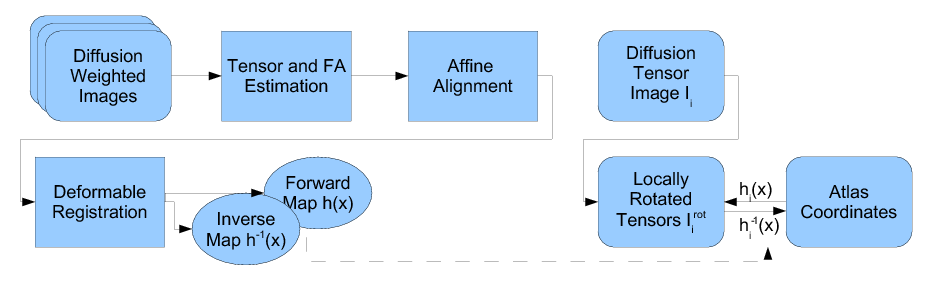
Our methodology for population analysis of DT-MRI is based on unbiased non-rigid registration of a population to a common coordinate system. The registration jointly produces an average DTI | Our methodology for population analysis of DT-MRI is based on unbiased non-rigid registration of a population to a common coordinate system. The registration jointly produces an average DTI | ||
| − | atlas, which is unbiased with respect to the choice of a template | + | atlas, which is unbiased with respect to the choice of a template image, along with diffeomorphic correspondence between each image. The |
| − | |||
registration image match metric uses a feature detector for thin fiber | registration image match metric uses a feature detector for thin fiber | ||
structures of white matter, and interpolation and averaging of diffusion | structures of white matter, and interpolation and averaging of diffusion | ||
| − | tensors use the Riemannian symmetric space framework. The | + | tensors use the Riemannian symmetric space framework. The anatomically significant correspondence provides a basis for comparison of tensor |
| − | |||
features and fiber tract geometry in clinical studies. | features and fiber tract geometry in clinical studies. | ||
| + | |||
| + | [[Image:goodlett_dti_atlas_flowchart.png]] | ||
| + | |||
| + | Our registration procedure is based on a scalar feature image which is sensitive to sheet like structures. We have observed that the major fiber bundles of interest occur as sheet or tube like manifolds in the FA image of the brain. As a feature image we use the maximum eigenvalue of the hessian of the FA image. | ||
| + | |||
| + | {| border="1" | ||
| + | |+'''Scalar feature image''' | ||
| + | |- | ||
| + | | [[Image:cbg-fa-axial.png|256px|Axial slice of FA image]]<br />Axial FA | ||
| + | | [[Image:cbg-fa-coronal.png|256px|Coronal slice of FA image]]<br />Coronal FA | ||
| + | | [[Image:cbg-fa-saggital.png|256px|Sagittal slice of FA image]]<br />Sagittal FA | ||
| + | |- | ||
| + | | [[Image:curv-axial.png|256px|Axial slice of feature image]]<br />Axial feature | ||
| + | | [[Image:curv-coronal.png|256px|Coronal slice of feature image]]<br />Coronal feature | ||
| + | | [[Image:curv-saggital.png|256px|Sagittal slice of feature image]]<br />Sagittal feature | ||
| + | |} | ||
= Publications = | = Publications = | ||
Revision as of 20:37, 2 April 2007
Home < Algorithm:UNC:DTI:Population AnalysisDescription
Our methodology for population analysis of DT-MRI is based on unbiased non-rigid registration of a population to a common coordinate system. The registration jointly produces an average DTI atlas, which is unbiased with respect to the choice of a template image, along with diffeomorphic correspondence between each image. The registration image match metric uses a feature detector for thin fiber structures of white matter, and interpolation and averaging of diffusion tensors use the Riemannian symmetric space framework. The anatomically significant correspondence provides a basis for comparison of tensor features and fiber tract geometry in clinical studies.
Our registration procedure is based on a scalar feature image which is sensitive to sheet like structures. We have observed that the major fiber bundles of interest occur as sheet or tube like manifolds in the FA image of the brain. As a feature image we use the maximum eigenvalue of the hessian of the FA image.
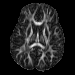 Axial FA |
 Coronal FA |
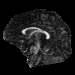 Sagittal FA |
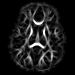 Axial feature |
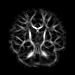 Coronal feature |
 Sagittal feature |
Publications
- Casey Goodlett, Brad Davis, Remi Jean, John Gilmore, Guido Gerig. Improved Correspondence for DTI Population Studies via Unbiased Atlas Building. Proc. MICCAI 2006, Springer LNCS v. 4191, pp. 260 - 267.PDF