Difference between revisions of "Algorithm:UNC:New"
| (33 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | Back to [[Algorithm:Main|NA-MIC Algorithms]] | |
| − | = | + | = Overview of UNC Algorithms = |
| − | |||
| − | + | At UNC, we are interested in a range of algorithms and solutions for the surface based analysis of brain structures and the cortex. We pioneered the use of spherical harmonics based shape analysis for comparing brain structures across objects. We are now working on incorporating various data sources on the entire cortical surface for improving the correspondence computation. | |
| − | + | = UNC Projects = | |
| − | + | {| | |
| − | [[ | + | | | [[Image:Sulcaldepth.png|thumb|left|200px]] |
| + | | | | ||
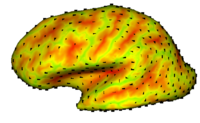
| − | == | + | == [[Projects:CorticalCorrespondenceWithParticleSystem|Cortical Correspondence using Particle System]] == |
| − | + | In this project, we want to compute cortical correspondence on populations, using various features such as cortical structure, DTI connectivity, vascular structure, and functional data (fMRI). This presents a challenge because of the highly convoluted surface of the cortex, as well as because of the different properties of the data features we want to incorporate together. [[Projects:CorticalCorrespondenceWithParticleSystem|More...]] | |
<font color="red">'''New: '''</font> | <font color="red">'''New: '''</font> | ||
| − | |||
| − | |||
| − | + | * The feature-based particle system implementation successfully works on toy data. | |
| + | * Currently testing with actual cortical data using sulcal depth. | ||
| + | |||
| + | |- | ||
| − | [[ | + | | | [[Image:UNCShape_OverviewAnalysis_MICCAI06.gif|thumb|left|200px]] |
| + | | | | ||
| − | == | + | == [[Projects:ShapeAnalysisFrameworkUsingSPHARMPDM|Shape Analysis Framework using SPHARM-PDM]] == |
| − | + | The UNC shape analysis is based on an analysis framework of objects with spherical topology, described mainly by sampled spherical harmonics SPHARM-PDM. The input of the shape analysis framework is a set of binary segmentations of a single brain structure, such as the hippocampus or caudate. These segmentations are converted into a shape description (SPHARM) with correspondence and analyzed via Hotelling T^2 two sample metric. [[Projects:ShapeAnalysisFrameworkUsingSPHARMPDM|More...]] | |
<font color="red">'''New: '''</font> | <font color="red">'''New: '''</font> | ||
| − | |||
| − | [ | + | * First version of Shape Analysis Toolset available as part of UNC Neurolib open source ([http://www.ia.unc.edu/dev/download/shapeAnalysis download]) , this is to be added to the NAMIC toolkit. |
| + | * Slicer 3 module for whole shape analysis pipeline in progress (based on BatchMake and distributed computing using Condor) | ||
| − | + | |- | |
| − | + | | | [[Image:UNCShape_ShapeCorrespondence.png|thumb|left|200px]] | |
| + | | | | ||
| − | + | == [[Projects:PopulationBasedCorrespondence|Population Based Correspondence]] == | |
| − | + | We are developing methodology to automatically find dense point correspondences between a collection of polygonal genus 0 meshes. The advantage of this method is independence from indivisual templates, as well as enhanced modeling properties. The method is based on minimizing a cost function that describes the goodness of correspondence. Apart from a cost function derived from the description length of the model, we also employ a cost function working with arbitrary local features. We extended the original methods to use surface curvature measurements, which are independent to differences of object aligment. [[Projects:PopulationBasedCorrespondence|More...]] | |
| − | + | <font color="red">'''New: '''</font> | |
| − | + | ||
| − | + | * Software available as part of UNC Neurolib open source ([http://www.ia.unc.edu/dev website]) | |
| + | * Evaluation on lateral ventricles, hippocampi, caudates, striatum, femural bone. Outperforms standard MDL on complex structures. | ||
| + | |||
| + | |- | ||
| + | |||
| + | | | [[Image:UNCShape_CaudatePval_MICCAI06.png|thumb|left|200px]] | ||
| + | | | | ||
| + | |||
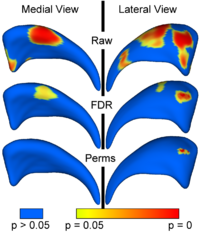
| + | == [[Projects:LocalStatisticalAnalysisViaPermutationTests|Local Statistical Analysis via Permutation Tests]] == | ||
| + | |||
| + | We have further developed a set of statistical testing methods that allow the analysis of local shape differences using the Hotelling T 2 two sample metric. Permutatioin tests are employed for the computation of statistical p-values, both raw and corrected for multiple comparisons. Resulting significance maps are easily visualized. Additional visualization of the group tests are provided via mean difference magnitude and vector maps, as well as maps of the group covariance information. Ongoing research focuses on incorporating covariates such as clinical scores into the testing scheme. [[Projects:LocalStatisticalAnalysisViaPermutationTests|More...]] | ||
<font color="red">'''New: '''</font> | <font color="red">'''New: '''</font> | ||
| − | * | + | * Available as part of Shape Analysis Toolset in UNC Neurolib open source ([http://www.ia.unc.edu/dev/download/shapeAnalysis download]). |
| − | + | ||
| − | + | |- | |
| − | + | | | [[Image:Cbg-dtiatlas-tracts.png|thumb|left|200px]] | |
| − | + | | | | |
| − | + | ||
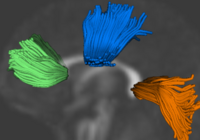
| − | + | == [[Projects:DTIPopulationAnalysis|Population Analysis from Deformable Registration]] == | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Analysis of populations of diffusion images typically requires time-consuming manual segmentation of structures of interest to obtain correspondance for statistics. This project uses non-rigid registration of DTI images to produce a common coordinate system for hypothesis testing of diffusion properties. [[Projects:DTIPopulationAnalysis|More...]] | |
| − | |||
| − | |||
| − | |||
| − | = | + | <font color="red">'''New: '''</font> Command line DTI tools available as part of UNC [http://www.ia.unc.edu/dev NeuroLib] |
| − | |||
| − | + | |- | |
| + | | style="width:10%" | [[Image:DTIQuantitativeAnalysis.png|thumb|left|200px]] | ||
| + | | style="width:90%" | | ||
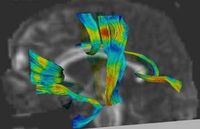
| − | + | == [[Projects:DTIQuantitativeTractAnalysis|Quantitative Analysis of Fiber Tract Bundles]] == | |
| − | |||
| − | |||
| − | |||
| − | + | DT-MRI tractography can be used as a coordinate system for computing statistics of diffusion tensor data. The quantitative analysis of diffusion tensors takes into account the space of tensor measurements using a nonlinear Riemannian symmetric space framework. Tracts of interest are represented as a medial spline attributed with cross-sectional statistics. [[Projects:DTIQuantitativeTractAnalysis|More...]] | |
| − | = | + | <font color="red">'''New: '''</font> Gilmore J, Lin W, Corouge I, Vetsa Y, Smith J, Kang C, Gu H, Hamer R, Lieberman J, Gerig G. Early Postnatal Development of Corpus Callosum and Corticospinal White Matter Assessed with Quantitative Tractography. AJNR Am J Neuroradiol. 2007. |
| − | + | |- | |
| − | + | | | [[Image:DTINoiseStatistics.png|thumb|left|200px]] | |
| + | | | | ||
| − | + | == [[Projects:DTINoiseStatistics|Influence of Imaging Noise on DTI Statistics]] == | |
| − | |||
| − | + | Clinical acquisition of diffusion weighted images with high signal to noise ratio remains a challenge. The goal of this project is to understand the impact of MR noise on quantiative statistics of diffusion properties such as anisotropy measures, trace, etc. [[Projects:DTINoiseStatistics|More...]] | |
| − | = | + | <font color="red">'''New: '''</font> Casey Goodlett, P. Thomas Fletcher, Weili Lin, Guido Gerig. Quantification of measurement error in DTI: Theoretical predictions and validation, MICCAI 2007. |
| − | + | |- | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | |} | |
| − | |||
Latest revision as of 22:22, 30 October 2007
Home < Algorithm:UNC:NewBack to NA-MIC Algorithms
Contents
- 1 Overview of UNC Algorithms
- 2 UNC Projects
- 2.1 Cortical Correspondence using Particle System
- 2.2 Shape Analysis Framework using SPHARM-PDM
- 2.3 Population Based Correspondence
- 2.4 Local Statistical Analysis via Permutation Tests
- 2.5 Population Analysis from Deformable Registration
- 2.6 Quantitative Analysis of Fiber Tract Bundles
- 2.7 Influence of Imaging Noise on DTI Statistics
Overview of UNC Algorithms
At UNC, we are interested in a range of algorithms and solutions for the surface based analysis of brain structures and the cortex. We pioneered the use of spherical harmonics based shape analysis for comparing brain structures across objects. We are now working on incorporating various data sources on the entire cortical surface for improving the correspondence computation.
UNC Projects
Cortical Correspondence using Particle SystemIn this project, we want to compute cortical correspondence on populations, using various features such as cortical structure, DTI connectivity, vascular structure, and functional data (fMRI). This presents a challenge because of the highly convoluted surface of the cortex, as well as because of the different properties of the data features we want to incorporate together. More... New:
| |
Shape Analysis Framework using SPHARM-PDMThe UNC shape analysis is based on an analysis framework of objects with spherical topology, described mainly by sampled spherical harmonics SPHARM-PDM. The input of the shape analysis framework is a set of binary segmentations of a single brain structure, such as the hippocampus or caudate. These segmentations are converted into a shape description (SPHARM) with correspondence and analyzed via Hotelling T^2 two sample metric. More... New:
| |
Population Based CorrespondenceWe are developing methodology to automatically find dense point correspondences between a collection of polygonal genus 0 meshes. The advantage of this method is independence from indivisual templates, as well as enhanced modeling properties. The method is based on minimizing a cost function that describes the goodness of correspondence. Apart from a cost function derived from the description length of the model, we also employ a cost function working with arbitrary local features. We extended the original methods to use surface curvature measurements, which are independent to differences of object aligment. More... New:
| |
Local Statistical Analysis via Permutation TestsWe have further developed a set of statistical testing methods that allow the analysis of local shape differences using the Hotelling T 2 two sample metric. Permutatioin tests are employed for the computation of statistical p-values, both raw and corrected for multiple comparisons. Resulting significance maps are easily visualized. Additional visualization of the group tests are provided via mean difference magnitude and vector maps, as well as maps of the group covariance information. Ongoing research focuses on incorporating covariates such as clinical scores into the testing scheme. More... New:
| |
Population Analysis from Deformable RegistrationAnalysis of populations of diffusion images typically requires time-consuming manual segmentation of structures of interest to obtain correspondance for statistics. This project uses non-rigid registration of DTI images to produce a common coordinate system for hypothesis testing of diffusion properties. More... New: Command line DTI tools available as part of UNC NeuroLib
| |
Quantitative Analysis of Fiber Tract BundlesDT-MRI tractography can be used as a coordinate system for computing statistics of diffusion tensor data. The quantitative analysis of diffusion tensors takes into account the space of tensor measurements using a nonlinear Riemannian symmetric space framework. Tracts of interest are represented as a medial spline attributed with cross-sectional statistics. More... New: Gilmore J, Lin W, Corouge I, Vetsa Y, Smith J, Kang C, Gu H, Hamer R, Lieberman J, Gerig G. Early Postnatal Development of Corpus Callosum and Corticospinal White Matter Assessed with Quantitative Tractography. AJNR Am J Neuroradiol. 2007. | |
Influence of Imaging Noise on DTI StatisticsClinical acquisition of diffusion weighted images with high signal to noise ratio remains a challenge. The goal of this project is to understand the impact of MR noise on quantiative statistics of diffusion properties such as anisotropy measures, trace, etc. More... New: Casey Goodlett, P. Thomas Fletcher, Weili Lin, Guido Gerig. Quantification of measurement error in DTI: Theoretical predictions and validation, MICCAI 2007. |