|
|
| Line 84: |
Line 84: |
| | * Clinical: | | * Clinical: |
| | ** Collaboration with [[DBP:Harvard|Harvard]] on shape analysis and DTI analysis. | | ** Collaboration with [[DBP:Harvard|Harvard]] on shape analysis and DTI analysis. |
| − |
| |
| − | = Old content, delete after full update =
| |
| − |
| |
| − | Nov 30, 2004
| |
| − |
| |
| − | == DTI: Quantitative analysis of fiber bundles ==
| |
| − |
| |
| − | === Components of FiberViewer Project ===
| |
| − |
| |
| − | Input: Sets of streamlines from tractography:
| |
| − | ITK polyline format with DTI attributes
| |
| − | Clustering of sets of streamlines to sets of
| |
| − | strong fiber bundles using various curve
| |
| − | distance metrics
| |
| − | Parametrization of sets of lines and
| |
| − | reparametrization for equidistant sampling
| |
| − | with arc-length
| |
| − | Calculation of local geometric properties of
| |
| − | streamlines (Frenet frame, curvature, torsion)
| |
| − | Attributing streamlines at each sample point
| |
| − | with DTI measurements (ADC, FA, lambda1..3,
| |
| − | ev. whole tensor) via trilinear interpolation
| |
| − | Calculation of DTI statistics within cross-
| |
| − | sections along fiber tracts after selection
| |
| − | of coordinate origin
| |
| − | Write statistics (DTI properties as a function
| |
| − | of arclength) to a text file for statistical
| |
| − | analysis.
| |
| − | Output: Processed, cleaned and clustered fiber
| |
| − | bundles (ITK polyline format) / Statistics
| |
| − | of DTI properties per selected bundle
| |
| − |
| |
| − | === Software ===
| |
| − |
| |
| − | * Algorithms written in ITK. GUI of prototype software written in QT ('''FiberViewer''' software). Prototype software tested in clinical studies at UNC. Validation tests with repeated DTI of same subject (6 cases).
| |
| − |
| |
| − | * Additionally available: ITK compatible fibertracking prototype tool '''FiberTracking''' to be used to study overlap/dissimilarity with other tools already available to NA-MIC: Functionality: reads raw MRI-DT data (6 direction Basser scheme), fiber tracking based on user-selected source and regions (S. Mori scheme), display of fibertracts and volumetric data, output: sets of streamlines in ITK polyline format attributedwith DTI properties and display parameteres (radiusof tubes, local color, etc.).
| |
| − |
| |
| − | ==== DTI Training Tools ====
| |
| − |
| |
| − | DTI Training Tools (Downloadable in zip package [[Image:DTI-Training-Tools.zip|Image:DTI-Training-Tools.zip]])
| |
| − |
| |
| − | * Glyphs - displays FA slices and tensor field
| |
| − | * Fiber - simplistic tractography from single voxel
| |
| − | * Conn - display of Riemannian flow from voxel
| |
| − | * [http://www.ia.unc.edu/dev/download/mriwatcher/index.htm MRIWatcher] - displays a set of MRI volumes simultaneously. MRTWatcher can also overlay segmentation mask.
| |
| − |
| |
| − | The manual for DTI Training Tools is included in zip file. Source code for the programs is available from [http://www.ia.unc.edu/dev UNC NeuroLib].
| |
| − |
| |
| − | === Recent Activities ===
| |
| − |
| |
| − | * UNC has developed a DTIFiberClass, which is now available as part of ITK
| |
| − | * UNC has initiated the discussion for a standard format within NAMIC of representing DTI Tensor data and DTI Fibers
| |
| − | * All software is available for download via anonymous download from our CVS server
| |
| − |
| |
| − | : pserver:anonymous@demeter.ia.unc.edu . Dashboard and Testing procedures have been installed and are operational.
| |
| − |
| |
| − | <br />
| |
| − |
| |
| − | === Plans ===
| |
| − |
| |
| − | * Feasibility tests on DTI data from NAMIC clinical partners
| |
| − | * DTI standardization issues (AHM SLC and follow-up)
| |
| − | * Evaluation of Slicer integration
| |
| − |
| |
| − | ==== Long term ====
| |
| − |
| |
| − | * DTI Fiber shape representation tools
| |
| − | * Correspondence via fiber bundles (shape, parametrization)
| |
| − | * Fiber clustering via Normalized Cuts (ITK filter)
| |
| − |
| |
| − | === Relationship to other NA-MIC partners ===
| |
| − |
| |
| − | DTI preprocessing (smoothing, interpolation):
| |
| − | MGH, Utah: Improve quality of raw DTI
| |
| − | data, Resampling DTI including tensor
| |
| − | re-orientation and full-tensor interpolation
| |
| − | DTI tensor statistics using Lie Group analysis:
| |
| − | Utah (Tom Fletcher), will replace the
| |
| − | simple averaging of FA, ADC, lambda1..3
| |
| − | as shown in "Averaging" step above
| |
| − | DTI tensor calculation, tractography, combined
| |
| − | display: Slicer, DoDTI (C.-F. Westin):
| |
| − | Processing of DTI to get fiber tracts of
| |
| − | interest for subsequent FiberViewer analysis
| |
| − | DTI atlas: probabilistic DTI atlas of normal
| |
| − | controls (MGH): To be used as reference
| |
| − | DTI atlas of normal controls
| |
| − | DTI annotated fiber tract atlas-template:
| |
| − | M. Farlow: To be used to calculate
| |
| − | fiber bundle properties for each annonated
| |
| − | tract, to serve as refrence template for
| |
| − | geometry and location of bundles as well
| |
| − | as for DTI properties as a function of
| |
| − | bundles.
| |
| − | DTI clinical data schizophrenia: M. Shenton
| |
UNC Algorithms page
Diffusion Tensor Imaging
Quantitative Analysis of Fiber Tract Bundles
DTI tractography can be used as a coordinate system for computing statistics of diffusion tensor data. The quantitative analysis of diffusion tensors takes into account the space of tensor measurements using a nonlinear Riemannian symmetric space framework. Tracts of interest are represented as a medial spline attributed with cross-sectional statistics.
More...
Description - Publications - Software
Population Analysis from Deformable Registration
Analysis of populations of diffusion images typically requires time-consuming manual segmentation of structures of interest to obtain correspondance for statistics. This project uses non-rigid registration of DTI images to produce a common coordinate system for hypothesis testing of diffusion properties.
More...
Description - Publications - Software
Shape Analysis of Brain Structures Across Groups
Shape analysis has become of increasing relevance to the neuroimaging community due to its potential to precisely locate morphological changes between healthy and pathological structures. This project focuses on developing novel methodology and a comprehensive set of tools for the computation of 3D structural statistical shape analysis. There are several open problems in this area, ranging from multi-object analysis, enhanced shape correspondence to statistical analysis of shape with clinical covariates.
UNC Shape Analysis Framework using SPHARM-PDM
|
The UNC shape analysis is based on an analysis framework of objects with spherical topology, described mainly by sampled spherical harmonics SPHARM-PDM. The input of the shape analysis framework is a set of binary segmentations of a single brain structure, such as the hippocampus or caudate. These segmentations are converted into a shape description (SPHARM) with correspondence and analyzed via Hotelling T^2 two sample metric. More...
New:
- First version of Shape Analysis Toolset available as part of UNC Neurolib open source (download) , this is to be added to the NAMIC toolkit.
- Toolset distribution contains open data for other researcher to evaluate novel shape analysis enhancements
- Slicer 3 modules for individual tools in the UNC Shape Analysis Toolset completed
- Slicer 3 module for whole shape analysis pipeline in progress (based on BatchMake and distributed computing using Condor)
- New shape analysis related papers:
- 2 submissions to the Main MICCAI 2007 conference
- Styner M, Xu SC, El-Sayed M, Gerig G, Correspondence Evaluation in Local Shape Analysis and Structural Subdivision, IEEE Symposium on Biomedical Imaging ISBI 2007, in print
- Zhou C, Park DC, Styner M, Wang YM, ROI Constrained Statistical Surface Morphometry, IEEE Symposium on Biomedical Imaging ISBI 2007, in print
- Nain D, Styner M, Niethammer M, Levitt JJ, Shenton ME, Gerig G, Bobick A, Tannenbaum A, Statistical Shape Analysis of Brain Structures Using Spherical Wavelets, IEEE Symposium on Biomedical Imaging ISBI 2007, in print
- M. Styner, I. Oguz, S. Xu, D. Pantazis, and G. Gerig. Statistical group differences in anatomical shape analysis using hotelling T2 metric. Proc SPIE Medical Imaging Conference, in print, 2007.
- C Cascio, M Styner, RG Smith, M Poe, G Gerig, H Hazlett, M Jomier, R Bammer, J Piven, Reduced relationship to cortical white matter revealed by tractography-based segmentation of the corpus callosum in yound children with developmental delay, Am J Psychiatry, 2006, (163) 2157-2163, December.
Description - Publications - Software
|
|
Population Based Correspondence
We are developing methodology to automatically find dense point correspondences between a collection of polygonal genus 0 meshes. The advantage of this method is independence from indivisual templates, as well as enhanced modeling properties. The method is based on minimizing a cost function that describes the goodness of correspondence. Apart from a cost function derived from the description length of the model, we also employ a cost function working with arbitrary local features. We extended the original methods to use surface curvature measurements, which are independent to differences of object aligment. More...
New:
- Software available as part of UNC Neurolib open source (website)
- Evaluation on lateral ventricles, hippocampi, caudates, striatum, femural bone. Outperforms standard MDL on complex structures.
- Submission to MICCAI 2007 conference
- Tobias Heimann, I. Oguz, I. Wolf, M. Styner, HP. Meinzer. Implementing the Automatic Generation of 3D Statistical Shape Models with ITK. Accepted to MICCAI 2006 Open Source Workshop. More...
Description - Publications - Software
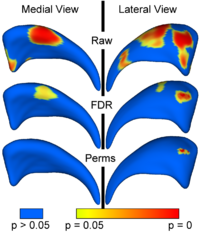
Local Statistical Analysis via Permutation Tests
We have further developed a set of statistical testing methods that allow the analysis of local shape differences using the Hotelling T 2 two sample metric. Permutatioin tests are employed for the computation of statistical p-values, both raw and corrected for multiple comparisons. Resulting significance maps are easily visualized. Additional visualization of the group tests are provided via mean difference magnitude and vector maps, as well as maps of the group covariance information. Ongoing research focuses on incorporating covariates such as clinical scores into the testing scheme. More...
New:
- Available as part of Shape Analysis Toolset in UNC Neurolib open source (download).
- M. Styner, I. Oguz, S. Xu, C. Brechbuehler, D. Pantazis, J. Levitt, M. Shenton, G. Gerig. Framework for the Statistical Shape Analysis of Brain Structures using SPHARM-PDM. Accepted to MICCAI 2006 Open Source Workshop. More...
Description - Publications - Software
Collaborations with other groups in NAMIC
- Algorithms:
- Shape Analysis
- Joint pipeline I/O formulation and development with Kitware (Brad Davis, Jim Miller) and MIT (Polina Golland)
- Use of UNC statistical analysis for spherical wavelet shape with GeorgiaTech (Delphine Nain) and Utah (Tom Fletcher)
- Use of UNC statistical analysis for combined multi-object correspondence establishment with Utah (Josh Cates, Tom Fletcher)
- DTI
- Statistics of tensors and noise in diffusion weighted imaging with Utah (Tom Fletcher)
- Clinical:
- Collaboration with Harvard on shape analysis and DTI analysis.