Difference between revisions of "Annotation Documentation"
| Line 168: | Line 168: | ||
Source Code: | Source Code: | ||
*[http://viewvc.slicer.org/viewcvs.cgi/trunk/QTModules/miAnnotation/ | *[http://viewvc.slicer.org/viewcvs.cgi/trunk/QTModules/miAnnotation/ | ||
| + | |||
| + | UML class diagrams of selected functionalities: | ||
| + | [[Image:Annotation Module TextNodes Class Diagram.png|thumb|left|Classes involved in adding Text annotations]] | ||
== More Information == | == More Information == | ||
Revision as of 16:12, 1 July 2010
Home < Annotation Documentation
miAnnotation
miAnnotation (Medical Image Annotation)
General Information
This module is a work in progress targeted for release with 3D Slicer version 4
Module Type & Category
Type: Interactive
Category: Base
Authors, Collaborators & Contact
- Kilian Pohl: pohl@csail.mit.edu
- Daniel Haehn: haehn@bwh.harvard.edu
- Past Contributors:
- Yong Zhang: yzhang@bwh.harvard.edu
Module Description
The miAnnotation Module is designed for users to annotate medical images to capture information latent in the scans.
Usage
Eight (8) different types of annotations can be created in the following ways:
- Region Of Interest (ROI): Click on the ROI Icon
 on the middle panel of the miAnnotation main panel, a new ROI annotation will then be created on the right-hand-side 3D view. There are seven (7) control points on the ROI annotation for the user to interact with. The six (6) control points located on the six surface of the ROI box can be dragged using mouse left-button to enlarge or shrink the box. The one (1) control point located in the center of the box can be dragged using mouse left-button to relocate the box in the 3D view. A new ROI entry will also be created in the annotation table. The properties of the ROI annotation can be edited by clicking the edit icon
on the middle panel of the miAnnotation main panel, a new ROI annotation will then be created on the right-hand-side 3D view. There are seven (7) control points on the ROI annotation for the user to interact with. The six (6) control points located on the six surface of the ROI box can be dragged using mouse left-button to enlarge or shrink the box. The one (1) control point located in the center of the box can be dragged using mouse left-button to relocate the box in the 3D view. A new ROI entry will also be created in the annotation table. The properties of the ROI annotation can be edited by clicking the edit icon  in the annotation table.
in the annotation table.
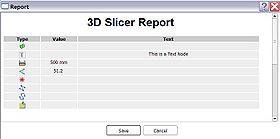
- Text: Click on the text Icon
 on the middle panel of the miAnnotation main panel, a new text annotation will then be created on the right-hand-side 3D view. The default location of the new text annotation is on the bottom left corner of the viewer. The user can move the text annotation to any other locations by clicking and dragging the text annotation using the mouse middle-button. A new text entry will also be created in the annotation table. The content of the text annotation can be edited from the entry in the annotation table. The properties of the text annotation can be edited by clicking the edit icon
on the middle panel of the miAnnotation main panel, a new text annotation will then be created on the right-hand-side 3D view. The default location of the new text annotation is on the bottom left corner of the viewer. The user can move the text annotation to any other locations by clicking and dragging the text annotation using the mouse middle-button. A new text entry will also be created in the annotation table. The content of the text annotation can be edited from the entry in the annotation table. The properties of the text annotation can be edited by clicking the edit icon  in the annotation table.
in the annotation table.
- Ruler: Click on the ruler Icon
 on the middle panel of the miAnnotation main panel, a new ruler annotation will then be created on the right-hand-side 3D view. The default location of the new ruler annotation is in the middle of the viewer spanned from left to right. The user can click and drag any one of the two control points of the ruler annotation to interact with it. A new ruler entry will also be created in the annotation table. The value of the ruler measurement can be automatically updated when the user interact with the ruler. The properties of the ruler annotation can be edited by clicking the edit icon
on the middle panel of the miAnnotation main panel, a new ruler annotation will then be created on the right-hand-side 3D view. The default location of the new ruler annotation is in the middle of the viewer spanned from left to right. The user can click and drag any one of the two control points of the ruler annotation to interact with it. A new ruler entry will also be created in the annotation table. The value of the ruler measurement can be automatically updated when the user interact with the ruler. The properties of the ruler annotation can be edited by clicking the edit icon  in the annotation table.
in the annotation table.
- Angle: Click on the angle Icon
 on the middle panel of the miAnnotation main panel, a new angle annotation will then be initialized on the right-hand-side 3D view. The users are required to manually click three (3) times in the 3D viewer using mouse left-button to create the real angle annotation. After creating the angle annotation, the user can click and drag any one of the three control points of the angle annotation to interact with it. A new angle entry will also be created in the annotation table. The value of the angle measurement can be automatically updated when the user interact with the angle. The properties of the annotation can be edited by clicking the edit icon
on the middle panel of the miAnnotation main panel, a new angle annotation will then be initialized on the right-hand-side 3D view. The users are required to manually click three (3) times in the 3D viewer using mouse left-button to create the real angle annotation. After creating the angle annotation, the user can click and drag any one of the three control points of the angle annotation to interact with it. A new angle entry will also be created in the annotation table. The value of the angle measurement can be automatically updated when the user interact with the angle. The properties of the annotation can be edited by clicking the edit icon  in the annotation table.
in the annotation table.
- Fiducial: Click on the fiducial Icon
 on the middle panel of the miAnnotation main panel, then the users are required to manually click in any of the 2D viewers using mouse left-button to create a new fiducial annotation. The new annotation will also be displayed in the 3D viewer. After creating the annotation, the user can click and drag it to interact with it. A new fiducial entry will also be created in the annotation table. The properties of the annotation can be edited by clicking the edit icon
on the middle panel of the miAnnotation main panel, then the users are required to manually click in any of the 2D viewers using mouse left-button to create a new fiducial annotation. The new annotation will also be displayed in the 3D viewer. After creating the annotation, the user can click and drag it to interact with it. A new fiducial entry will also be created in the annotation table. The properties of the annotation can be edited by clicking the edit icon  in the annotation table.
in the annotation table.
- Bidimensional Lines: Click on the bidimensional lines Icon
 on the middle panel of the miAnnotation main panel, then the users are required to manually click in the 3D viewers using mouse left-button to create two lines. These two lines are automatically orthogonal. After creating the annotation, the user can click and drag any of the four control points to interact with it. A new bidimensional line entry will also be created in the annotation table. The properties of the annotation can be edited by clicking the edit icon
on the middle panel of the miAnnotation main panel, then the users are required to manually click in the 3D viewers using mouse left-button to create two lines. These two lines are automatically orthogonal. After creating the annotation, the user can click and drag any of the four control points to interact with it. A new bidimensional line entry will also be created in the annotation table. The properties of the annotation can be edited by clicking the edit icon  in the annotation table.
in the annotation table.
- Spline: Click on the spline Icon
 on the middle panel of the miAnnotation main panel, then a new spline annotation point is created right in the center of the 3d viewer. The users are able to manually click and drag the spline point using mouse left-button to other locations in the 3D viewer. Up to five (5) points can be clicked and dragged from the initial location of the spline annotation. After creating the annotation, the user can click and drag any of the five control points to form different splines to fit their needs. Or the user can click and drag the entire spline annotation to move it as a whole. A new spline entry will also be created in the annotation table. The properties of the annotation can be edited by clicking the edit icon
on the middle panel of the miAnnotation main panel, then a new spline annotation point is created right in the center of the 3d viewer. The users are able to manually click and drag the spline point using mouse left-button to other locations in the 3D viewer. Up to five (5) points can be clicked and dragged from the initial location of the spline annotation. After creating the annotation, the user can click and drag any of the five control points to form different splines to fit their needs. Or the user can click and drag the entire spline annotation to move it as a whole. A new spline entry will also be created in the annotation table. The properties of the annotation can be edited by clicking the edit icon  in the annotation table.
in the annotation table.
- Sticky Note: Click on the stick note Icon
 on the middle panel of the miAnnotation main panel, then a new stick note is created. There are no displays of the note in any of the 2D or 3D viewers. After creating the annotation, a new sticky note entry will be created in the annotation table. The properties of the annotation can be edited by clicking the edit icon
on the middle panel of the miAnnotation main panel, then a new stick note is created. There are no displays of the note in any of the 2D or 3D viewers. After creating the annotation, a new sticky note entry will be created in the annotation table. The properties of the annotation can be edited by clicking the edit icon  in the annotation table.
in the annotation table.
Use Cases, Examples
This module is especially appropriate for these use cases:
- Use Case 1: This module is useful for basic measurements of volume, area, length, angle.
- Use Case 2: Use eight different annotations to annotate medical images.
Tutorials
Links to tutorials explaining how to use this module:
- Tutorial (coming soon)
- Data Set (coming soon)
Quick Tour of Features and Use
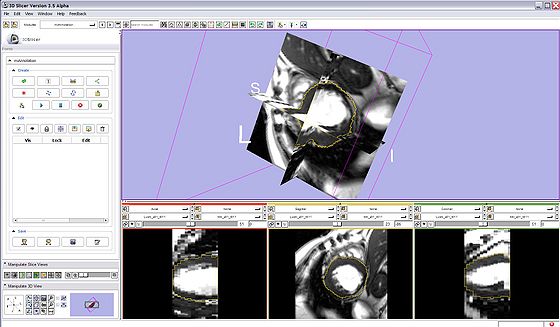
- miAnnotation Main Panel:
|
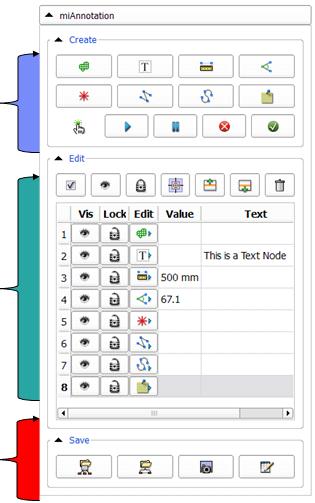
- Edit annotation table:
- Save Annotations:
|
Development
Notes from the Developer(s)
miAnnotation is not part of Slicer3.6. To use the module, you have to use the trunk version of the Slicer3. See the build instructions.
Dependencies
Fiducials and Measurements Modules are currently needed. Later these will be integrated into a unified Annotations module.
Tests
On the Dashboard, these tests verify that the module is working on various platforms:
- vtkSlicermiAnnotationModuleLogicTest1.cxx
- vtkMRMLAnnotationStorageNodeTest1.cxx
- vtkMRMLAnnotationRulerStorageNodeTest1.cxx
- vtkMRMLAnnotationRulerNodeTest1.cxx
- vtkMRMLAnnotationPointDisplayNodeTest1.cxx
- vtkMRMLAnnotationNodeTest1.cxx
- vtkMRMLAnnotationLinesStorageNodeTest1.cxx
- vtkMRMLAnnotationLinesNodeTest1.cxx
- vtkMRMLAnnotationLineDisplayNodeTest1.cxx
- vtkMRMLAnnotationFiducialsStorageNodeTest1.cxx
- vtkMRMLAnnotationFiducialNodeTest1.cxx
- vtkMRMLAnnotationDisplayNodeTest1.cxx
- vtkMRMLAnnotationControlPointsStorageNodeTest1.cxx
- vtkMRMLAnnotationControlPointsNodeTest1.cxx
- vtkMRMLAnnotationAngleStorageNodeTest1.cxx
- vtkMRMLAnnotationAngleNodeTest1.cxx
- qSlicermiAnnotationModuleAnnotationPropertyDialogTest1.cxx
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Source Code:
UML class diagrams of selected functionalities:
More Information
Acknowledgment
This work is supported by an ARRA supplement to the Neuroimage Analysis Center (NAC), an NCRR Resource Center, NIH NCRR grant P41 RR13218. This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005419.