Difference between revisions of "CTSC:ARRA.011210"
From NAMIC Wiki
| (9 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
'''Agenda''' | '''Agenda''' | ||
| − | + | i2b2 to XNAT to DICOM architecture and workflow | |
== Harvard Catalyst Medical Informatics group Meeting Minutes January 12, 2010 == | == Harvard Catalyst Medical Informatics group Meeting Minutes January 12, 2010 == | ||
| Line 26: | Line 26: | ||
'''Discussion''' | '''Discussion''' | ||
| − | + | ===Project components=== | |
| − | + | ||
| − | + | * the software development part. | |
| − | + | * the department engagement (Randy is working on setting up meetings with the different institutions to work on policy issues). | |
| + | * the studies: we need the science, it is essential to get a couple of publications to validate the project. | ||
<br> | <br> | ||
| Line 37: | Line 38: | ||
[[image:mi2b2.png]] | [[image:mi2b2.png]] | ||
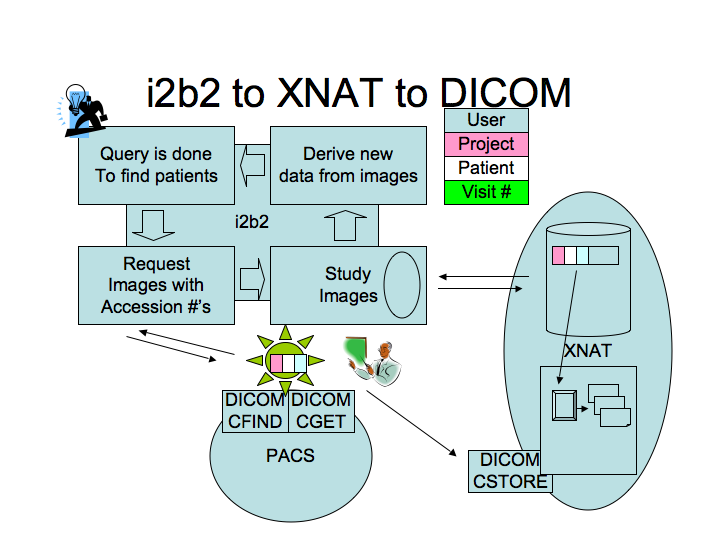
| − | *In the picture above, it is clear now that | + | *In the picture above, it is clear now that the software to be developed by this project, represented by the STAR icon in the figure, will be an intermediary between the PACS and XNAT. i2b2 will not communicate directly with the PACS. Anything built in i2b2 will communicate with STAR and then STAR will communicate with the PACS. |
| − | * i2b2 structure is | + | * i2b2 structure is organized by projects and users. Every message from i2b2 always includes who the user is and information about the project (IRB etc). We have to understand how to coordinate the user/project information structure from i2b2 with that of XNAT. Randy mentioned that Dan Marcus has just been asked to support communication between i2b2 and XNAT for another project at Emory University. |
| − | * | + | * STAR will need to be able to log into XNAT and send the images to the right place. The most difficult part will probably be to send back the new data from XNAT to i2b2. |
| − | * patient: in i2b2 there is a structure to ensure that the access | + | * patient: in i2b2 there is a structure to ensure that the access to the HIPAA data is limited. There is a data management cell in i2b2 to deal with the medical record number (MRN). STAR will need the MRN to retrieve data. |
| − | * How is XNAT prepared to deal with | + | * How is XNAT prepared to deal with MRNs? According to Yong, XNAT does not allow you to store any HIPAA controlled data. |
| + | * Each project will be formed around an IRB, then around a set of patients, then a set of users allowed to use the data associated with the project. | ||
| + | <br> | ||
| + | |||
| + | === i2b2 to XNAT to DICOM workflow=== | ||
| + | |||
| + | *Request is made from i2b2 client to look up: | ||
| + | **By accession number: What are the nature of accessions for "studies" and "images" at the 4 sites? | ||
| + | **By patient: Assumed to always be a MRN | ||
| + | ** i2b2 enforces that the PI is asking only for the studies he is allowed to see. | ||
| + | ** As we discussed last week, the accession # is mapped to the UID (very internal, one per image, they are part of the DICOM). | ||
<br> | <br> | ||
| − | + | *A navigational client experience is made to identify a study or an image: | |
| + | ** How can we give access to only a couple of studies and not the whole PACS? Answer: The STAR software will serve as a gatekeeper and the throttle to limit access both in terms of when it can retrieve images and what images to retrieve (e.g. images from the last 5 years are OK, older no because it takes too long to get access and download). | ||
| − | * | + | <br> |
| − | ** | + | *Hints are made as to what images are useful: |
| − | ** | + | ** Would this be possible? |
| − | * | + | ** The description is not always the best place to look for hint but for example one could look for bolus injection to make sure contrast agent was used. |
| − | ** | + | |
| + | <br> | ||
| + | * Transfer is specified (to be discussed at next meeting) | ||
| + | ** Target location | ||
| + | ** Set up XNAT to receive image | ||
| + | |||
| + | <br> | ||
| + | *Request is queued (to be discussed at next meeting) | ||
| + | **Log of transfer is available | ||
| + | **Notification that transfer is complete is made | ||
Latest revision as of 15:54, 19 January 2010
Home < CTSC:ARRA.011210Back to CTSC:ARRA supplement
Agenda
i2b2 to XNAT to DICOM architecture and workflow
Contents
Harvard Catalyst Medical Informatics group Meeting Minutes January 12, 2010
In attendance:
- Valerie Humblet
- Mike Mendis
- Shawn Murphy
- Bill Tellier
- Mark Anderson
- Randy Gollub
- Yong Gao
- Paul Lamonica
- Steve Piper
- Wendy Plesniak
- Alex Zeitsev
Discussion
Project components
- the software development part.
- the department engagement (Randy is working on setting up meetings with the different institutions to work on policy issues).
- the studies: we need the science, it is essential to get a couple of publications to validate the project.
i2b2 to XNAT to DICOM architecture
- In the picture above, it is clear now that the software to be developed by this project, represented by the STAR icon in the figure, will be an intermediary between the PACS and XNAT. i2b2 will not communicate directly with the PACS. Anything built in i2b2 will communicate with STAR and then STAR will communicate with the PACS.
- i2b2 structure is organized by projects and users. Every message from i2b2 always includes who the user is and information about the project (IRB etc). We have to understand how to coordinate the user/project information structure from i2b2 with that of XNAT. Randy mentioned that Dan Marcus has just been asked to support communication between i2b2 and XNAT for another project at Emory University.
- STAR will need to be able to log into XNAT and send the images to the right place. The most difficult part will probably be to send back the new data from XNAT to i2b2.
- patient: in i2b2 there is a structure to ensure that the access to the HIPAA data is limited. There is a data management cell in i2b2 to deal with the medical record number (MRN). STAR will need the MRN to retrieve data.
- How is XNAT prepared to deal with MRNs? According to Yong, XNAT does not allow you to store any HIPAA controlled data.
- Each project will be formed around an IRB, then around a set of patients, then a set of users allowed to use the data associated with the project.
i2b2 to XNAT to DICOM workflow
- Request is made from i2b2 client to look up:
- By accession number: What are the nature of accessions for "studies" and "images" at the 4 sites?
- By patient: Assumed to always be a MRN
- i2b2 enforces that the PI is asking only for the studies he is allowed to see.
- As we discussed last week, the accession # is mapped to the UID (very internal, one per image, they are part of the DICOM).
- A navigational client experience is made to identify a study or an image:
- How can we give access to only a couple of studies and not the whole PACS? Answer: The STAR software will serve as a gatekeeper and the throttle to limit access both in terms of when it can retrieve images and what images to retrieve (e.g. images from the last 5 years are OK, older no because it takes too long to get access and download).
- Hints are made as to what images are useful:
- Would this be possible?
- The description is not always the best place to look for hint but for example one could look for bolus injection to make sure contrast agent was used.
- Transfer is specified (to be discussed at next meeting)
- Target location
- Set up XNAT to receive image
- Request is queued (to be discussed at next meeting)
- Log of transfer is available
- Notification that transfer is complete is made