CTSC:ARRA.121509
From NAMIC Wiki
Revision as of 05:21, 19 January 2010 by RandyGollub (talk | contribs) (→Harvard Catalyst Medical Informatics group Meeting Minutes December 15, 2009)
Home < CTSC:ARRA.121509
Back to CTSC:ARRA supplement
Agenda
- DICOM query/retrieve tool
Harvard Catalyst Medical Informatics group Meeting Minutes December 15, 2009
In attendance:
- Valerie Humblet
- Yong Gao
- Mike Mendis
- Randy Gollub
- Mark Anderson
- Steve Piper
- Karl Helmer
- Alex Zeitsev
- Shawn Murphy
- Jesse Wei
- Paul Lamonica
- Bill Tellier
1. Welcome
- The mi2b2 group welcomed Paul Lamonica (Senior PACS analyst) and Bill Tellier (PACS manager), both from CHB.
2. DICOM query/retreive tool
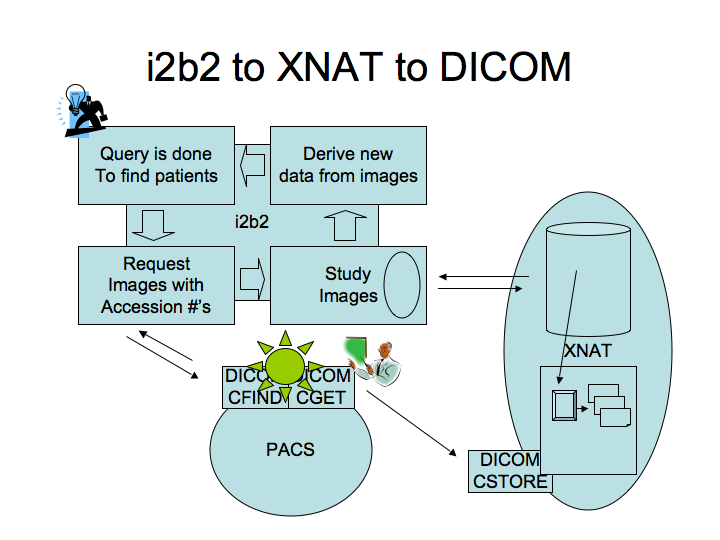
Here is a schema of the project, the DICOM query/retrieve tool is represented by the yellow STAR.
- Shawn mentioned that one key problem to solve is to understand how the users will be restricted from viewing everything for each patient. Departments are concerned about the time limit during which a PI has access to the data, the scope of what they can see, how an audit trail can be maintained, who to contact to create a new user. Note: HIPAA laws will applied to these data because they were acquired under clinical practice.
- We also have to decide how STAR does know to deliver data to the investigator. STAR will be told to go to the PACS and get data. Only the people with an IRB approval will have access, they will need an authorized account. With the help of i2b2 project management, STAR will have to check that it can retrieve data from PACS. It includes a username, token (to make sure the communication is legitimate).
- i2b2 and STAR should be coordinated. Any time STAR gets a request, it confirms that it comes from i2b2 and is authorized. STAR is a relatively passive system for the most part. It will have some permission to get DICOM data from the PACS and then send it to XNAT or another repository.
- i2b2 and STAR will use XML messages to communicate.
- STAR will be administered by an administrator. IT at each institution will be able to monitor everything but will not have the responsibilities to write the code. There is a need for a collaborative research effort to do it.
- Steve reminded the group that individual PIs in the Radiology Departments already have the kind of permission we want to implement with STAR. They get access to their data and load them on their own computer.
- How large of a data request can be handled without bringing down the PACS? The biggest problem will be for archived data at institutions that use archival systems with slow access times. At those institutions there will be a big difference between recent data and archived data. To address this concern, the team must learn about the behavior of the different PACS systems.
- Some PACS, such as GE at BIDMC, have different priority levels for access and actions. At CHB, they have Synapse that does not have variable priority levels, everything is online.
- At CHB they are in the process of setting up a research PACS.
- Some conclusions: three questions emerged from this meeting:
- How do we assure that only the authorized people get access to the data requested?
- How do we assure that we don't bring down the system?
- How do we set up a process to interrogate the PACS to make sure the data are usable for the study?
- Jesse said that one will need to be able to view at least 1 image from the series. In academics, depending if they are more research oriented or not, people tend to modify the protocols. It is different in the private practice.
- Steve says that the curation process of the data should be the PI's task.