Difference between revisions of "Collaboration/Iowa/Meshing/Migrate Iowa Neural Net code to pure ITK"
From NAMIC Wiki
| (7 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
| − | + | {| | |
| + | |[[Image:ProjectWeek-2007.png|thumb|320px|Return to [[2007_Programming/Project_Week_MIT|Project Week Main Page]] ]] | ||
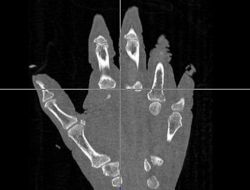
| + | |[[Image:Hand-Atlas.jpg|thumb|left|250px|Atlas Hand Image]] | ||
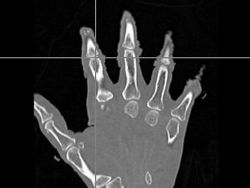
| + | |[[Image:Hand-Subject.jpg|thumb|left|250px|Subject Hand Image]] | ||
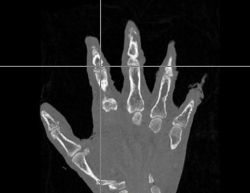
| + | |[[Image:Hand-WarpedAtlas.jpg|thumb|left|250px|Warped Atlas Image]] | ||
| + | |} | ||
| − | = | + | |
| − | + | ||
| + | |||
| + | |||
| + | __NOTOC__ | ||
| + | ===Key Investigators=== | ||
| + | * Iowa: Vincent Magnotta and Nicole Grosland | ||
| + | * Kitware: Stephen Alyward | ||
| + | |||
| + | <div style="margin: 20px;"> | ||
| + | |||
| + | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
| + | |||
| + | <h1>Objective</h1> | ||
| + | Conversion of the Iowa Neural netwok segmentation using a flexible module for image registration supporting all ITK transforms written using the transform I/O in ITK. Conversion to the ITK neural network from the current annie implementation may also be | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
| + | |||
| + | <h1>Approach, Plan</h1> | ||
| + | |||
| + | Integrate additional ITK transform types into the neural network using the general ITK transform I/O mechanism. Evaluate the changes to segment the phalanx bones bones of the hand. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div style="width: 40%; float: left;"> | ||
| + | |||
| + | <h1>Progress</h1> | ||
| + | Two milestones were reached in this effort. | ||
| + | |||
| + | *The first was that we have integrated a rigid body initialization for a Thirion Demons registration for atlas <-> subject registration. The above modification was used to warp the atlas to the subject as shown in the figures. | ||
| + | |||
| + | *The second was that the neural network code now supports all ITK transforms for registration of the atlas<->subject. This was achieved using the itkTransformReader and the a templated function over the image type and transform type to resample the images. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <br style="clear: both;" /> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | ===References=== | ||
| + | * Magnotta VA, Heckel D, Andreasen NC, Cizadlo T, Corson PW, Ehrhardt JC, Yuh WT. "Measurement of brain structures with artificial neural networks: two- and three-dimensional applications", Radiology 211 (1999), 781-90. | ||
| + | * Powell S, Magnotta VA, Johnson HJ, Jammalamadaka VK, Andreasen NC. "Registration and Machine Learning Based Automated Segmentation of Subcortical and Cerebellar Brain Structures", NeuroImage, In Press. | ||
Latest revision as of 02:59, 29 June 2007
Home < Collaboration < Iowa < Meshing < Migrate Iowa Neural Net code to pure ITK Return to Project Week Main Page |
Key Investigators
- Iowa: Vincent Magnotta and Nicole Grosland
- Kitware: Stephen Alyward
Objective
Conversion of the Iowa Neural netwok segmentation using a flexible module for image registration supporting all ITK transforms written using the transform I/O in ITK. Conversion to the ITK neural network from the current annie implementation may also be
Approach, Plan
Integrate additional ITK transform types into the neural network using the general ITK transform I/O mechanism. Evaluate the changes to segment the phalanx bones bones of the hand.
Progress
Two milestones were reached in this effort.
- The first was that we have integrated a rigid body initialization for a Thirion Demons registration for atlas <-> subject registration. The above modification was used to warp the atlas to the subject as shown in the figures.
- The second was that the neural network code now supports all ITK transforms for registration of the atlas<->subject. This was achieved using the itkTransformReader and the a templated function over the image type and transform type to resample the images.
References
- Magnotta VA, Heckel D, Andreasen NC, Cizadlo T, Corson PW, Ehrhardt JC, Yuh WT. "Measurement of brain structures with artificial neural networks: two- and three-dimensional applications", Radiology 211 (1999), 781-90.
- Powell S, Magnotta VA, Johnson HJ, Jammalamadaka VK, Andreasen NC. "Registration and Machine Learning Based Automated Segmentation of Subcortical and Cerebellar Brain Structures", NeuroImage, In Press.