Difference between revisions of "Collaboration:NCMIR Microscopy"
From NAMIC Wiki
| Line 15: | Line 15: | ||
* [[media:Microscopy-Confocal-TrainingTutorial-2009JUNE.pdf|Confocal Microscopy Tutorial]] | * [[media:Microscopy-Confocal-TrainingTutorial-2009JUNE.pdf|Confocal Microscopy Tutorial]] | ||
| − | + | ==Grant#== | |
| + | U54EB005149-05S3 | ||
| + | ==Key Personnel== | ||
| + | Bryan Smith, PI, Jeffrey S. Grethe | ||
| + | ==Grant Duration== | ||
== Contact == | == Contact == | ||
* Mark Ellisman (mark at ncmir.ucsd.edu) | * Mark Ellisman (mark at ncmir.ucsd.edu) | ||
* Jeffrey Grethe (jgrethe at ncmir.ucsd.edu) | * Jeffrey Grethe (jgrethe at ncmir.ucsd.edu) | ||
Revision as of 20:18, 30 November 2009
Home < Collaboration:NCMIR MicroscopyContents
Background
- The collaboration in regards to Microscopy was fostered through a supplement, funding now completed, to the National Center for Microscopy and Imaging Research (NCMIR; http://ncmir.ucsd.edu) at the University of California, San Diego. A key emphasis of NCMIR is the application of advanced imaging technologies to the nervous system in health and disease. Affiliated with UCSD’s Center for Research in Biological Systems (CRBS), the NCMIR is a recognized authority in the development of technologies for high throughput multi-scale imaging and analysis of biological systems at the mesoscale, the dimensional range spanning 5 nm3 and 50µm3. Macromolecules, organelles, and multi-component structures like synapses which are encompassed in this dimensional range have traditionally been challenging to study because they fall in the resolution gap between X-ray crystallography at one end and medical imaging at the other.
- Work, as part of this collaboration, focused on requirements and extensions for the Slicer environment to address multi-scale microscopy imaging data, thereby allowing for the analysis and visualization of cellular data. The project has focused on the importation of data into Slicer and the investigation of algorithms for cellular and sub-cellular data (e.g. see - http://www.na-mic.org/Wiki/index.php/Projects/Slicer3/2007_Project_Week_Support_for_electron_microscopy).
Delivered Using Slicer for Microscopy Tutorial
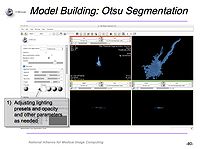
- A direct result of this collaboration was the tutorial developed by UCSD for use of Slicer with confocal microscopy data: "Guiding you step by step through the process of loading confocal microscopy data, working with that data, and creating a 3D model for visualization." The tutorial covers the complete use of Slicer from loading data (including the downloading of data from publicly available open-source repositories), visualization of data and basic editing with the editor module. The tutorial then covers model building using basic slicer modules and the Otsu segmentation module.
- This tutorial won the 2009 Summer Programming Week Tutorial contest (see - http://www.na-mic.org/Wiki/index.php/Events:TutorialContestJune2009).
Grant#
U54EB005149-05S3
Key Personnel
Bryan Smith, PI, Jeffrey S. Grethe
Grant Duration
Contact
- Mark Ellisman (mark at ncmir.ucsd.edu)
- Jeffrey Grethe (jgrethe at ncmir.ucsd.edu)