Difference between revisions of "DBP1:Irvine"
| (9 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | Back to [[DBP1:Main|NA-MIC DBP 1]] | |
| + | __NOTOC__ | ||
| + | = Overview of UC Irvine DBP 1 = | ||
| − | + | Researchers at UCI investigate the connections between neuroanatomy and schizophrenia. | |
| − | + | For more introductory information, follow this [[DBP:Irvine|link]]. | |
= UC Irvine Projects = | = UC Irvine Projects = | ||
| − | {| | + | {| cellpadding="10" style="text-align:left;" |
| − | | style="width: | + | |
| − | | style="width: | + | | style="width:15%" | [[Image:Dlpfc1.jpg|200px]] |
| + | | style="width:85%" | | ||
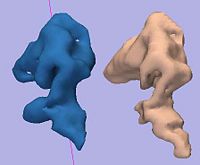
| − | == [[Projects: | + | == [[Projects:RuleBasedDLPFCSegmentation|Rule-Based DLPFC Segmentation]] == |
| − | In this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules | + | In this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the dorsolateral prefrontal cortex. [[Projects:RuleBasedDLPFCSegmentation|More...]] |
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> Al-Hakim, et al. A Dorsolateral Prefrontal Cortex Semi-Automatic Segmenter. SPIE MI 2006. |
|- | |- | ||
| − | | | [[Image: | + | | | [[Image:MBIRNseedROIcc1.png|200px]] |
| | | | | | ||
| Line 26: | Line 29: | ||
To carry out quantitative and qualitative validation of the DTI tractography tools. These will be applied to a limited set of specific tracts in single data sets and single tractography tools, and on several data sets using at least two tractography programs and by investigators in different laboratories. [[Projects:DTIValidation|More...]] | To carry out quantitative and qualitative validation of the DTI tractography tools. These will be applied to a limited set of specific tracts in single data sets and single tractography tools, and on several data sets using at least two tractography programs and by investigators in different laboratories. [[Projects:DTIValidation|More...]] | ||
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> Future research is needed to establish critical values for diffusion sequence acquisition parameters that would allow diffusion data processing via Slicer. |
|- | |- | ||
| − | | | | + | | | |
| | | | | | ||
| Line 37: | Line 40: | ||
In a legacy dataset of 28 chronic schizophrenics, to determine fMRI activation during working memory in schizophrenia and apply novel tools to assess circuitry between regions through combining behavior and BOLD signal analyses. [[Projects:ImagingPhenotypesInSchizophrenicsAndControls|More...]] | In a legacy dataset of 28 chronic schizophrenics, to determine fMRI activation during working memory in schizophrenia and apply novel tools to assess circuitry between regions through combining behavior and BOLD signal analyses. [[Projects:ImagingPhenotypesInSchizophrenicsAndControls|More...]] | ||
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> The initial results were presented at the Human Brain Mapping meeting in Florence IT 2006. |
|- | |- | ||
| − | | | | + | | | |
| | | | | | ||
| Line 48: | Line 51: | ||
To use structural equation modeling to compare attentional circuitry within the legacy set of 28 schizophrenic subjects who performed similar attentional tasks in both a PET and fMRI scan. [[Projects:AttentionalCircuitsInSchizophreniaInfMRIAndPET|More...]] | To use structural equation modeling to compare attentional circuitry within the legacy set of 28 schizophrenic subjects who performed similar attentional tasks in both a PET and fMRI scan. [[Projects:AttentionalCircuitsInSchizophreniaInfMRIAndPET|More...]] | ||
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> The data can be found via the NAMIC website. |
|- | |- | ||
| − | | | | + | | | |
| | | | | | ||
| Line 59: | Line 62: | ||
To compare morphometric/volumetric differences in a multi-site anatomical sample of schizophrenic subjects, in collaboration with the FBIRN consortium. Subject morphometric scans will be sent to UCSD's morphometric processing lab (Dale) for automated processing. A system for submitting morphometric scans to processing pipeline and retreiving/wrapping the results for inclusion into FBIRN HIDs will be developed. The project will also be used to drive the development of the workflow processing system being developed by Shawn Murphey's group. [[Projects:MultiSiteMorhometry|More...]] | To compare morphometric/volumetric differences in a multi-site anatomical sample of schizophrenic subjects, in collaboration with the FBIRN consortium. Subject morphometric scans will be sent to UCSD's morphometric processing lab (Dale) for automated processing. A system for submitting morphometric scans to processing pipeline and retreiving/wrapping the results for inclusion into FBIRN HIDs will be developed. The project will also be used to drive the development of the workflow processing system being developed by Shawn Murphey's group. [[Projects:MultiSiteMorhometry|More...]] | ||
| − | <font color="red">'''New: '''</font> | + | <font color="red">'''New: '''</font> Two hundred subjects have been collected, in some cases with a repeated T1 scan, in 3T and 1.5T scanners. Discussions of morphometric analysis methods are underway. |
|} | |} | ||
Latest revision as of 15:30, 21 August 2009
Home < DBP1:IrvineBack to NA-MIC DBP 1
Overview of UC Irvine DBP 1
Researchers at UCI investigate the connections between neuroanatomy and schizophrenia.
For more introductory information, follow this link.
UC Irvine Projects
|
Rule-Based DLPFC SegmentationIn this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the dorsolateral prefrontal cortex. More... New: Al-Hakim, et al. A Dorsolateral Prefrontal Cortex Semi-Automatic Segmenter. SPIE MI 2006. |
|
DTI ValidationTo carry out quantitative and qualitative validation of the DTI tractography tools. These will be applied to a limited set of specific tracts in single data sets and single tractography tools, and on several data sets using at least two tractography programs and by investigators in different laboratories. More... New: Future research is needed to establish critical values for diffusion sequence acquisition parameters that would allow diffusion data processing via Slicer. |
Optimal Imaging Phenotypes In Schizophrenics and ControlsIn a legacy dataset of 28 chronic schizophrenics, to determine fMRI activation during working memory in schizophrenia and apply novel tools to assess circuitry between regions through combining behavior and BOLD signal analyses. More... New: The initial results were presented at the Human Brain Mapping meeting in Florence IT 2006. | |
Attentional Circuits In Schizophrenia In fMRI And PETTo use structural equation modeling to compare attentional circuitry within the legacy set of 28 schizophrenic subjects who performed similar attentional tasks in both a PET and fMRI scan. More... New: The data can be found via the NAMIC website. | |
MultiSite MorphometryTo compare morphometric/volumetric differences in a multi-site anatomical sample of schizophrenic subjects, in collaboration with the FBIRN consortium. Subject morphometric scans will be sent to UCSD's morphometric processing lab (Dale) for automated processing. A system for submitting morphometric scans to processing pipeline and retreiving/wrapping the results for inclusion into FBIRN HIDs will be developed. The project will also be used to drive the development of the workflow processing system being developed by Shawn Murphey's group. More... New: Two hundred subjects have been collected, in some cases with a repeated T1 scan, in 3T and 1.5T scanners. Discussions of morphometric analysis methods are underway. |