Difference between revisions of "DBP2:Queens:Roadmap"
Sidd queens (talk | contribs) |
Sidd queens (talk | contribs) |
||
| Line 13: | Line 13: | ||
*'''Tutorial''': Documentation will be written for a tutorial and sample data sets will be provided to perform simulated biopsies. | *'''Tutorial''': Documentation will be written for a tutorial and sample data sets will be provided to perform simulated biopsies. | ||
| − | + | == '''Roadmap''' == | |
| − | The primary goal for the roadmap is to develop an interventional module for Slicer3 for MRI-guided prostate biopsies. This module and the accompanying tutorial will serve as a template for interventional applications with Slicer3. The module will provide the necessary functionality for calibrating the robot to the MR scanner, planning biopsies, computing the necessary robot trajectory to perform each biopsy, and verification via post-biopsy images. | + | The primary goal for the roadmap is to develop an interventional module for Slicer3 for MRI-guided prostate biopsies. This module and the accompanying tutorial will serve as a template for interventional applications with Slicer3. The module will provide the necessary functionality for calibrating the robot to the MR scanner, planning biopsies, computing the necessary robot trajectory to perform each biopsy, and verification via post-biopsy images. We will obtain a biopsy plan from multi-parametric endorectal image volumes, executable with an existing prostate biopsy device. The system will be will be implemented under Slicer3 as an interactive application. |
<center> | <center> | ||
| Line 30: | Line 30: | ||
</center> | </center> | ||
| − | |||
| − | |||
| − | |||
=== '''Current Status of System Components and schedule of progress''' === | === '''Current Status of System Components and schedule of progress''' === | ||
Revision as of 15:38, 29 April 2009
Home < DBP2:Queens:RoadmapBack to NA-MIC Internal Collaborations, JHU DBP 2
Objective
We would like to create an end-to-end application within the NA-MIC Kit to enable an existing transrectal prostate biopsy device to perform multi-parametric MRI guided prostate biopsy in closed-bore high-field MRI magnets.
This page describes the technology roadmap for robotic prostate biopsy in the NA-MIC Kit. The basic components necessary for this application are:
- Tissue segmentation: Should be multi-modality, correcting for intensity inhomogeneity and work for both supine and prone patients, all imaged with an endorectal coil (ERC).
- Registration: co-registration of MRI datasets taken at different times, in different body positions, and under different imaging parameters
- Prostate Measurement: Measure volume of all segmented structures
- Biopsy Device Parameters: Geometry, kinematics, and calibration/registration of the robot system must be available in some form. This capability is not currently part of the NA-MIC kit. The application will be modular, to enable use of different devices.
- Tutorial: Documentation will be written for a tutorial and sample data sets will be provided to perform simulated biopsies.
Roadmap
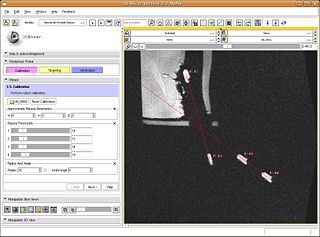
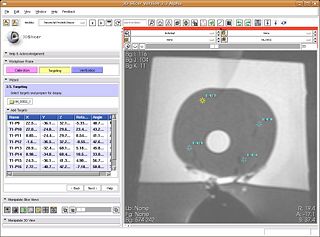
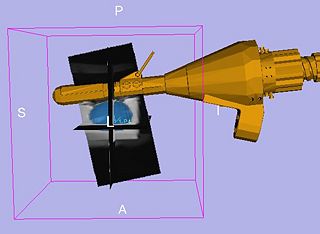
The primary goal for the roadmap is to develop an interventional module for Slicer3 for MRI-guided prostate biopsies. This module and the accompanying tutorial will serve as a template for interventional applications with Slicer3. The module will provide the necessary functionality for calibrating the robot to the MR scanner, planning biopsies, computing the necessary robot trajectory to perform each biopsy, and verification via post-biopsy images. We will obtain a biopsy plan from multi-parametric endorectal image volumes, executable with an existing prostate biopsy device. The system will be will be implemented under Slicer3 as an interactive application.
Current Status of System Components and schedule of progress
Software & documentation
- The TRProstateBiopsy module is in the "Queens" directory of the NAMICSandBox - access online
Tutorial (end-to-end):
- Trans-rectal prosate biopsy module: TransRectalProstateBiopsyTutorial
- Trans-rectal prostate biopsy tutorial dataset: TRPBTutorialDataset.zip
Publications
- Krieger A, Susil RC, Menard C, Coleman JA, Fichtinger G, Atalar E, Whitcomb LL, Design of A Novel MRI Compatible Manipulator for Image Guided Prostate Intervention, IEEE Trans. Biomed. Eng. 2005; 52(2):306-313
- Susil RC, Ménard C, Krieger A, Coleman JA, Camphausen K, Choyke P, Ullman K, Smith S, Fichtinger G, Whitcomb LL, Coleman NC, Atalar E, Transrectal Prostate Biopsy and Fiducial Marker Placement in a Standard 1.5T MRI Scanner, J Urol. 2006 Jan;175(1):113-20
- GS Fischer, A Krieger, I Iordachita, LL Whitcomb, and G Fichtinger, "MRI Compatibility of Robot Actuation Techniques -- A Comparative Study", Eleventh International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Proceedings in Lecture Notes in Computer Science Vol. 5242, pp 509-517, Springer, 2008
- Siddharth Vikal, Steven Haker, Clare Tempany, Gabor Fichtinger, Prostate contouring in MRI guided biopsy, SPIE Medical Imaging 2009
- S. Gill, P Abolmaesumi, S Vikal, P Mousavi, G. Fichtinger, Intraoperative Prostate Tracking with Slice-to-Volume Registration in MRI, 20th International Conference of the Society for Medical Innovation and Technology (SMIT), Vienna, Austria, August 28-31, 2008, Electronic proceedings, ISBN 3-902087-25-0, pp. 154-158
- Mewes PW, Tokuda J, DiMaio SP, Fischer GS, Csoma C, Gobbi DG, Tempany CM, Fichtinger G, Hata N, An Integrated MRI and Robot Control Software for an MRI compatible Robot in Prostate Intervention, International Conference on Robotics and Automation - ICRA 2008, Pasadena, CA, pp 2950-2962, May 2008
Team and Institutes
- PI: Gabor Fichtinger, Queen’s University (gabor at cs.queensu.ca)
- Co-I: Purang Abolmaesumi, Queen’s University (purang at cs.queensu.ca)
- Software Engineer Lead: Siddharth Vikal, Queen’s University (vikal at cs.queensu.ca
- Software Engineer: David Gobbi, Queen’s University (dgobbi at cs.queensu.ca)
- JHU Software Engineer Support: Csaba Csoma, Johns Hopkins University, csoma at jhu.edu
- NA-MIC Engineering Contact: Katie Hayes, MSc, Brigham and Women's Hospital, hayes at bwh.harvard.edu
- NA-MIC Algorithms Contact: Allen Tannenbaum, PhD, GeorgiaTech, tannenba at ece.gatech.edu
- Host Institutes: Queen's University & Johns Hopkins University