Difference between revisions of "DBP2:UNC:Regional Cortical Thickness Pipeline"
From NAMIC Wiki
| Line 4: | Line 4: | ||
<center> | <center> | ||
{| | {| | ||
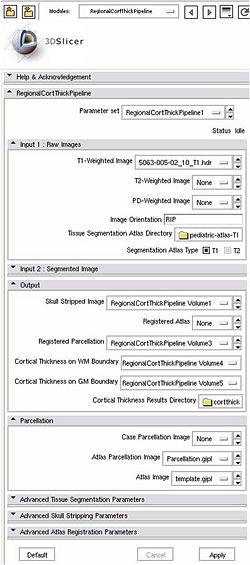
| − | |[[Image: | + | |[[Image:Screenshot_Slicer.jpg|thumb|250px|Screenshot of the application]] |
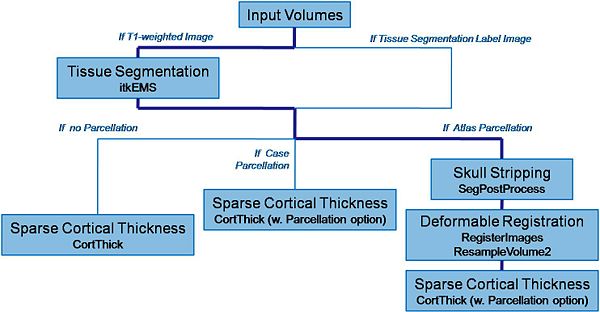
| − | |valign="center"|[[Image: | + | |valign="center"|[[Image:Schema_pipeline.jpg|thumb|600px|Pipeline description]] |
| − | |||
| − | |||
|} | |} | ||
</center> | </center> | ||
| Line 69: | Line 67: | ||
:* 1 normal control | :* 1 normal control | ||
| + | <center> | ||
| + | {| | ||
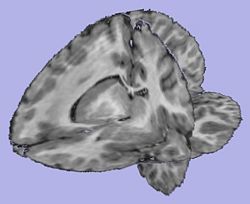
| + | |[[Image:T1Image.jpg|thumb|250px|T1-weighted skull-stripped image]] | ||
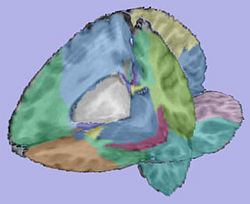
| + | |valign="center"|[[Image:Parcellation.jpg|thumb|250px|Parcellation image]] | ||
| + | |valign="center"|[[Image:WMThickness.jpg|thumb|250px|Cortical thickness on WM surface]] | ||
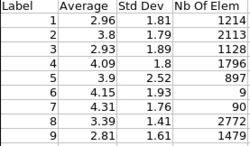
| + | |valign="center"|[[Image:ThicknessInformation.jpg|thumb|250px|Cortical thickness information]] | ||
| + | |} | ||
| + | </center> | ||
=== In progress === | === In progress === | ||
Revision as of 20:32, 17 October 2008
Home < DBP2:UNC:Regional Cortical Thickness PipelineBack to UNC Cortical Thickness Roadmap
Objective
We would like to create an end-to-end application within Slicer3 allowing individual and group analysis of regional cortical thickness.
General information
Pipeline description (steps)
Input: T1-weighted image, T2-weighted image, PD-weighted image
- 1. Tissue segmentation
- Tool: itkEMS (UNC Slicer3 external module)
- 2. Skull stripping
- Tool: SegPostProcess (UNC Slicer3 external module)
- 3. Deformable registration of pediatric regional atlas
- 3.1 Deformable registration of T1-weighted pediatric atlas
- Tool: RegisterImages (Slicer3 module)
- 3.2. Applying transformation to the parcellation map
- Tool: ResampleVolume2 (Slicer3 module)
- 3.1 Deformable registration of T1-weighted pediatric atlas
- 4. Cortical Thickness
- Tool: CortThick (UNC Slicer3 module)
- 1. Tissue segmentation
All the tools used in the current pipeline are Slicer3 modules, some of them being UNC external modules. The user can thus compute a regional cortical thickness analysis on one data, either within Slicer3 or by running the tools as command lines.
Usage (Command Line)
Inputs: T1-weighted image, T1-weigthed atlas, regional atlas (parcellation map)
- 1. Tissue segmentation
- Input: EMSparam.xml
- Output: Image_Corrected_EMS.gipl, Label.gipl
- 1. Tissue segmentation
itkEMSCLP --XMLFile EMSparam.xml
- 2. Skull stripping
- Input: Label.gipl, Image_Corrected_EMS.gipl
- Output: Image_Corrected_EMS_Stripped.gipl, BinaryMask.gipl (optional)
- 2. Skull stripping
SegPostProcessCLP Label.gipl Image_Corrected_EMS_Stripped.gipl --skullstripping Image_Corrected_EMS.gipl (--mask BinaryMask.gipl –dilate)
- 3. Deformable registration of pediatric regional atlas
- 3.1 Deformable registration of T1-weighted pediatric atlas
- Input: Atlas.gipl, Image_Corrected_EMS_Stripped.gipl
- Output: Atlas_Registered.gipl, Atlas_Registered_Transform.txt
- 3.1 Deformable registration of T1-weighted pediatric atlas
- 3. Deformable registration of pediatric regional atlas
RegisterImages Image_Corrected_EMS_Stripped.gipl Atlas.gipl –resampledImage Atlas_Registered.gipl –saveTransform Atlas_Registered_Transform.txt –registration PipelineBSpline
- 3.2. Applying transformation to the parcellation map
- Input: Parcellation.gipl, Atlas_Registered_Transform.txt, Image_Corrected_EMS_Stripped.gipl
- Output: Parcellation_Registered.gipl
- 3.2. Applying transformation to the parcellation map
ResampleVolume2 Parcellation.gipl Parcellation_Registered.gipl -f Atlas_Registered_Transform.txt -i nn -R Image_Corrected_EMS_Stripped.gipl
- 4. Cortical Thickness
- Input: Parcellation_Registered.gipl, Label.gipl
- Output: CortThick_Result_Dir/, WMCorticalThicknessMap, GMCorticalThicknessMap
- 4. Cortical Thickness
CortThickCLP CortThick_Result_Dir/ --par Parcellation_Registered.gipl --inputSeg Label.gipl --SaveWM WMCorticalThicknessMap --SaveGM GMCorticalThicknessMap
Analysis on a small pediatric dataset
Tests have been computed on a small pediatric dataset, including 2 years old and 4 years old cases
- 2 Autistic cases
- 1 developmental delay
- 1 normal control
In progress
- Workflow for individual analysis (Slicer3 external module using BatchMake)
Future work
- UNC Slicer3 external modules available on NITRIC
- Pediatric atlas (T1-weighted image and parcellation map) available to the community (XNAT?)
- Workflow for group analysis