Difference between revisions of "DBP2:UNC:Tissue Segmentation Evaluation"
(New page: Back to NA-MIC Internal Collaborations, UNC DBP 2 __NOTOC__ == Objective == We would like to evaluate the brain tissue segmentation using ...) |
|||
| (9 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | Back to [[ | + | Back to [[DBP2:UNC:Cortical_Thickness_Roadmap | UNC Cortical Thickness Roadmap]], [[DBP2:UNC:Regional_Cortical_Thickness_Pipeline | UNC Regional Cortical Thickness Pipeline]] |
__NOTOC__ | __NOTOC__ | ||
| Line 14: | Line 14: | ||
A UNC dataset has been used for this study, one patient being scanned 10 times on the same scanner. | A UNC dataset has been used for this study, one patient being scanned 10 times on the same scanner. | ||
Two dual-channel segmentation trials were performed to compare the segmentation reliability using the two algorithms. | Two dual-channel segmentation trials were performed to compare the segmentation reliability using the two algorithms. | ||
| + | A single T1-weighted image was registered to the UNC template atlas and the rest of the T1 images were affinely registered and scaled to that original T1 image to minimize scanner-influenced differences. | ||
=== Results === | === Results === | ||
| Line 19: | Line 20: | ||
One case failed at the execution using EMSegment and has been removed. | One case failed at the execution using EMSegment and has been removed. | ||
| − | The covariance was computed | + | The covariance was computed for each trial, not only considering each tissue (White Matter, Grey Matter and CSF), but also the Intra Cranial Volume, sum of the three tissues. |
- Covariance/Tissue WM GM CSF ICV | - Covariance/Tissue WM GM CSF ICV | ||
| Line 25: | Line 26: | ||
- EMSegment 7.66% 3.15% 14.84% 1.51% | - EMSegment 7.66% 3.15% 14.84% 1.51% | ||
| − | See results detailed on spreadsheet | + | See results detailed on [[Media:UNCDBP2_TissueSegmentationReliabilityStudy.xls | spreadsheet]]. |
| + | |||
| + | For each algorithm and each tissue, an average T1-weighted label image has been computed. Then, using the 'EvaluateSegmentationResult' tool, volume differences, average distances and tanimoto errors have been computed between each case and the previously computed average. | ||
| + | |||
| + | See data detailed on [[Media:UNCDBP2_TissueSegmentationReliabilityStudy2.xls | spreadsheet]]. | ||
| + | |||
| + | === Screenshots === | ||
| + | |||
| + | <center> | ||
| + | {| | ||
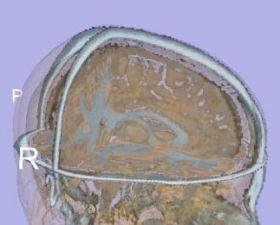
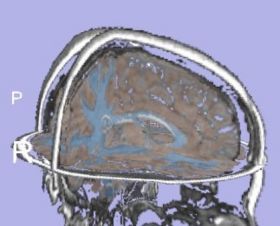
| + | |[[Image:TissueSegmentation_Adult_EMSegment.jpg|thumb|280px|EMSegment tissue segmentation on adult case]] | ||
| + | |valign="top"|[[Image:Image-TissueSegmentation_Adult_itkEMS.jpg|thumb|280px|itkEMS tissue segmentation on adult case]] | ||
| + | |} | ||
| + | </center> | ||
== Qualitative study on pediatric dataset == | == Qualitative study on pediatric dataset == | ||
| + | |||
| + | <center> | ||
| + | {| | ||
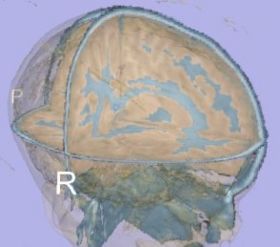
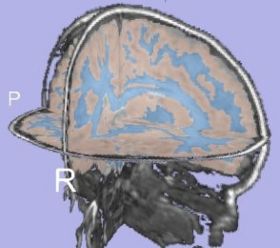
| + | |[[Image:TissueSegmentation_Pediatric_EMSegment.jpg|thumb|280px|EMSegment tissue segmentation on pediatric case]] | ||
| + | |valign="top"|[[Image:TissueSegmentation_Pediatric_itkEMS.jpg|thumb|280px|itkEMS tissue segmentation on pediatric case]] | ||
| + | |} | ||
| + | </center> | ||
Latest revision as of 19:17, 27 August 2010
Home < DBP2:UNC:Tissue Segmentation EvaluationBack to UNC Cortical Thickness Roadmap, UNC Regional Cortical Thickness Pipeline
Objective
We would like to evaluate the brain tissue segmentation using two different algorithms: itkEMS and Slicer3 EMSegment.
- A reliability study was performed on adult dataset.
- A qualitative study was performed on pediatric dataset.
Reliability study on adult dataset
Method
A UNC dataset has been used for this study, one patient being scanned 10 times on the same scanner. Two dual-channel segmentation trials were performed to compare the segmentation reliability using the two algorithms. A single T1-weighted image was registered to the UNC template atlas and the rest of the T1 images were affinely registered and scaled to that original T1 image to minimize scanner-influenced differences.
Results
One case failed at the execution using EMSegment and has been removed.
The covariance was computed for each trial, not only considering each tissue (White Matter, Grey Matter and CSF), but also the Intra Cranial Volume, sum of the three tissues.
- Covariance/Tissue WM GM CSF ICV - itkEMS 1.27% 0.99% 6.27% 0.88% - EMSegment 7.66% 3.15% 14.84% 1.51%
See results detailed on spreadsheet.
For each algorithm and each tissue, an average T1-weighted label image has been computed. Then, using the 'EvaluateSegmentationResult' tool, volume differences, average distances and tanimoto errors have been computed between each case and the previously computed average.
See data detailed on spreadsheet.