Difference between revisions of "DBP3:UCLA"
| Line 317: | Line 317: | ||
==Proceedings== | ==Proceedings== | ||
| + | * Bo Wang, Marcel W. Prastawa, Suyash P. Awate, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. Van Horn and Guido Gerig (2012) Segmentation of serial MRI of TBI patients using personalized atlas construction and topological change estimation Proceedings of the Tenth International Symposium on Biomedical Imaging (ISBI 2012), May 2-5, 2012, Barcelona, Catalonia, Spain | ||
* Bo Wang, Marcel W. Prastawa, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. van Horn, Guido Gerig (2012) A patient-tailored segmentation framework for longitudinal MR images of traumatic brain injury Proceedings of the Twenty-Fifth International Conference of SPIE—The International Society for Optical Engineering, February 4-9, 2012, San Diego, California, USA | * Bo Wang, Marcel W. Prastawa, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. van Horn, Guido Gerig (2012) A patient-tailored segmentation framework for longitudinal MR images of traumatic brain injury Proceedings of the Twenty-Fifth International Conference of SPIE—The International Society for Optical Engineering, February 4-9, 2012, San Diego, California, USA | ||
* Marc Niethammer, Gabriel L. Hart, Danielle F. Pace, Micah C. Chambers, Andrei Irimia, John D. van Horn, Stephen R. Aylward (2011) Geometric metamorphosis Proceedings of the Fourteenth International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2011), September 18-22, 2011, Toronto, Ontario, Canada | * Marc Niethammer, Gabriel L. Hart, Danielle F. Pace, Micah C. Chambers, Andrei Irimia, John D. van Horn, Stephen R. Aylward (2011) Geometric metamorphosis Proceedings of the Fourteenth International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2011), September 18-22, 2011, Toronto, Ontario, Canada | ||
Revision as of 04:44, 3 December 2011
Home < DBP3:UCLABack to NA-MIC DBPs | NA-MIC Cores
Contents
- 1 Introduction
- 2 Project Goals and Specific Aims
- 3 Methods
- 4 Current Tasks
- 5 Results
- 6 Outreach
- 6.1 Outreach Events
- 6.1.1 UCLA Neurology Science Day (January 2011)
- 6.1.2 TBI DBP Meeting at NA-MIC AHM (January 2011)
- 6.1.3 TBI DBP Conference Call with Utah Team (September 9, 2011)
- 6.1.4 NA-MIC Conference Call (October 27, 2011)
- 6.1.5 TBI DBP Conference Call (November 10, 2011)
- 6.1.6 NA-MIC Conference Call (November 16, 2011)
- 6.1 Outreach Events
- 7 Investigators
- 8 Awards and Honors
- 9 Publications
- 10 Links
- 11 Literature
Introduction
What is traumatic brain injury?
Traumatic brain injury, often referred to as "TBI", is most often an acute event that results in severe damage to portions of the brain. TBI results when the head suddenly and violently hits an object, or when an object pierces the skull and enters brain tissue. Symptoms of can be mild, moderate, or severe, depending on the extent of the damage to the brain. Common disabilities include cognitive deficits, sensory processing, communication, and behavior or mental health. Severe TBI may result in stupor where an individual can be aroused briefly by a strong stimulus (e.g. sharp pain); coma, where an individual is totally unconscious, unresponsive, unaware, and un-arousable; vegetative state, where an individual is unconscious and unaware of his or her surroundings, but continues to have a sleep-wake cycle and periods of alertness; and a persistent vegetative state, where an individual remains unresponsive for more than a month.
According to the CDC (United States Centers for Disease Control and Prevention), there are approximately 1.7 million people in the U.S. who suffer from a traumatic brain injury each year. 50,000 people die from TBI each year and 85,000 people suffer long term disabilities. In the U.S., more than 5.3 million people live with disabilities caused by TBI. Patients admitted to a hospital for TBI are included in this count, while those treated in an emergency room or doctor's office are not counted. The causes of TBI are diverse. The top three causes are: car accident, firearms and falls. Firearm injuries are often fatal: 9 out of 10 people die from such injuries. Young adults and the elderly are the age groups at highest risk for TBI. Along with a traumatic brain injury, persons are also susceptible to spinal cord injuries which is another type of traumatic injury that can result out of vehicle crashes, firearms and falls. Prevention of TBI is the best approach since there is no cure or way to reverse brain damage of this kind.
Mechanisms of TBI
Understanding the various mechanisms of TBI can be helpful for the development of robust and reliable computational algorithms for neuroimage data processing. These mechanisms are the highest causes of brain injury: Open head Injury, Closed Head Injury, Deceleration Injuries, Chemical/Toxic, Hypoxia, Tumors, Infections and Stroke.
1. Open Head Injury
* Results from bullet wounds, etc. * Largely focal damage * Penetration of the skull * Effects can be just as serious as closed brain injury
2. Closed Head Injury
* Resulting from a slip and fall, motor vehicle crashes, etc. * Focal damage and diffuse damage to axons * Effects tend to be broad (diffuse) * No penetration to the skull
3. Deceleration Injuries (Diffuse Axonal Injury) The skull is hard and inflexible while the brain is soft with the consistency of gelatin. The brain is encased inside the skull. During the movement of the skull through space (acceleration) and the rapid discontinuation of this action when the skull meets a stationary object (deceleration) causes the brain to move inside the skull. The brain moves at a different rate than the skull because it is soft. Different parts of the brain move at different speeds because of their relative lightness or heaviness. The differential movement of the skull and the brain when the head is struck results in direct brain injury, due to diffuse axonal shearing, contusion and brain swelling.
Diffuse axonal shearing: when the brain is slammed back and forth inside the skull it is alternately compressed and stretched because of the gelatinous consistency. The long, fragile axons of the neurons (single nerve cells in the brain and spinal cord) are also compressed and stretched. If the impact is strong enough, axons can be stretched until they are torn. This is called axonal shearing. When this happens, the neuron dies. After a severe brain injury, there is massive axonal shearing and neuron death.
4. Chemical/Toxic
* Also known as metabolic disorders * This occurs when harmful chemicals damage the neurons * Chemicals and toxins, e.g. insecticides, solvents, carbon monoxide poisoning, lead poisoning, etc.
5. Hypoxia (Lack of Oxygen)
* If the blood flow is depleted of oxygen, then irreversible brain injury can occur from anoxia (no oxygen) or hypoxia (reduced oxygen) * This condition may be caused by heart attacks, respiratory failure, drops in blood pressure and a low oxygen environment * This type of brain injury can result in severe cognitive and memory deficits
6. Tumors
* Tumors caused by cancer can grow on or over the brain * Tumors can cause brain injury by invading the spaces of the brain and causing direct damage * Damage can also result from pressure effects around an enlarged tumor * Surgical procedures to remove the tumor may also contribute to brain injury
7. Infections
* The brain and surrounding membranes are very prone to infections if the special blood-brain protective system is breached * Viruses and bacteria can cause serious and life-threatening diseases of the brain (encephalitis) and meninges (meningitis)
8. Stroke
* If blood flow is blocked through a cerebral vascular accident (stroke), cell death in the area deprived of blood will result * If there is bleeding in or over the brain (hemorrhage or hematoma) because of a tear in an artery or vein, loss of blood flow and injury to the brain tissue by the blood will also result in brain damage
In this DBP, we will be primarily concentrating on #'s 1, 2, and 3.
Neurological Concomitants of TBI
Following TBI, a cascade of neuroanatomical alterations initiate, with diffuse alterations in cortical structure peripheral to the point of injury but also distributed throughout the brain. Notably, there is ventricular enlargement and cortical thickness changes remote from the site of the TBI. White matter connectivity can be significantly altered with greatly reduced efficiency of signal transduction over affected pathways or complete cessation of inter-regional communication due to axonal damage. This can have profound effects on speech, motor, and cognitive processes. The extent of change is putatively related to the severity of TBI, location, subject age, and post-injury treatment, among other factors.
Challenges of TBI Neuroimaging
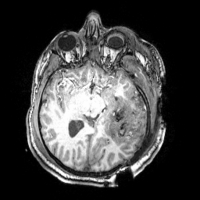
On anatomical MRI scans, TBI-related insults can appear as hyper-intensities, varying in magnitude and extent the degree to which tends to correlate with clinical symptoms. Additionally, in severe TBI, sections of skull, fractured during the injury or removed during surgical intervention, may not form a contiguous boundary enabling efficient digital removal of bone and other non-brain tissues, complicating tissue segmentation, regional parcellation, the measurement of ventricular size, cortical thickness, and other metrics. Computational algorithms require refinement to include constraints to account for TBI related signal alterations in anatomical scans; e.g. users may have to manually indicate regions encompassing the site of injury on the scans to guide local processing around the site and to reduce the weight of these regions on other, non-affected brain areas. Alternatively, probabilistic classifiers may need to include an extra classification for voxels whose tissue properties have been altered by TBI. This is particularly the case in diffusion weighted imaging (DWI) where the presence of TBI-related alterations in signal may reflect specific damage to white matter proximal to the lesion as well as long reaching effects along tracts to peripherally connected regions of cortex.
Project Goals and Specific Aims
Project Goals
An increasingly relevant means for the neurological assessment of traumatic brain injury (TBI) is with in vivo neuroimaging. However, standard automated image analysis methods are not robust with respect to the TBI-related changes in image contrast, changes in brain shape, cranial fractures, white matter fiber alterations, and other signatures of head injury. Multimodal quantification of brain insults and associating these with clinical and outcome metrics is a particular challenge. The emphasis in this DBP is placed on the feasibility of subject-specific analysis, as opposed to population-based averaging, to examine the influence of TBI on time-dependent alteration of gray and white matter integrity with accompanying change in clinical outcome variables to be used in subsequent TBI assessment. This DBP [1] seeks to:
1. Develop end-to-end processing approaches using the NA-MIC Kit to investigate alterations in cortical thickness, and subsequent ventricular and white matter changes in patients with TBI and in age-matched controls. Image processing will include segmentation of lesions, hemorrhage, edema, and other pathology relevant to TBI. Longitudinal changes will be assessed by registration and joint segmentation of baseline and follow-up data.
2. Develop robust workflows for diffusion weighted imaging (e.g. DTI, HARDI) datasets from TBI patients, by using the NA-MIC Kit and Slicer to obtain reliable and robust metrics of white matter pathology and of white matter changes due to therapy and/or recovery.
3. Using the NA-MIC Kit, cross-correlate multimodal metrics of cortical thickness, complexity, ventricular volume, and lesions from structural imaging and white matter fiber integrity from diffusion tensor imaging, with clinical outcome variables, i.e., time since injury, age, gender and other potential factors predictive of recovery.
Specific Aims
In this NA-MIC DBP, we seek to develop the means for guided and semi-automatic TBI analysis and quantification with a view toward assessing clinical improvement under the following Specific Aims:
Aim 1
How can multimodal assessment of altered brain anatomy speak directly to questions of brain plasticity and to secondary neuroanatomical effects of TBI?
We will develop end-to-end processing approaches using the NA-MIC Kit to investigate alterations in cortical thickness, subsequent ventricular, and white matter changes in patients with TBI and in age-matched controls.
Aim 2
Can multimodal workflows be developed to guide clinicians in the analysis and display of white matter fiber tract pathology that frequently accompanies brain insult?
In diffusion weighted imaging (e.g. DTI, HARDI) datasets from TBI patients, we will develop robust workflows using the NA-MIC Kit and Slicer to obtain reliable and robust metrics of white matter pathology.
Aim 3
How can multimodal metrics of TBI grey and white matter pathology be utilized to inform and guide clinical assessment?
Under this DBP, using sophisticated NA-MIC tools, we will develop end-to-end processing solutions by which to examine TBI neuroimaging data. The NA-MIC Kit encompasses a collection of tools for automated or semi-automated processing of medical imaging data. Notable is the Insight Toolkit (http://www.itk.org) for use in brain registration and segmentation via the 3D Slicer (http://www.slicer.org) software platform. These software tools may be linked to form data processing workflows that can process data via end-to-end solutions that may be shared with others, posted on websites, and used in training materials. They form an excellent platform for user-guided, patient-specific analysis, however, require additional development to inform the program about regions where TBI-related signal changes may necessitate alteration of model parameters or search volumes. Using the NA-MIC Kit, multimodal metrics of cortical thickness, complexity, and ventricular-volume from structural imaging and white matter fiber integrity from diffusion tensor imaging will be cross-correlated with clinical outcome variables, time since injury, age, gender and other potential factors predictive of recovery. We will emphasize the feasibility of subject-specific analysis, as opposed to population-based averaging, to examine the influence of TBI on time-dependent alteration of gray and white matter integrity with accompanying change in clinical outcome variables to be used in subsequent TBI assessment.
This DBP directly pertains to stated scientific and funding objectives of the NIBIB and other NIH institutes conducting (e.g. NINDS) and supporting (e.g. NCRR) research on the mechanisms underlying CNS injury; to develop intervention strategies to limit the primary and secondary brain damage occurring within days of a TBI; and to devise therapies to treat TBI and help in long-term recovery of function. DBP results will have important implications for national health policy concerning TBI awareness, treatment and brain plasticity. Thus, this DBP is directly in-line with the NIH mission for greater assessment of neurological insults and factors that predict long-term outcomes in individual patients.
Methods
Software and Analysis Protocol
We will specifically adopt the NA-MIC kit open source software platform consisting of Slicer, tools and toolkits such as VTK and ITK, and software engineering methodologies for multiplatform implementation. Using ITK, data will be intensity normalized and bias-field corrected; tissue types will be segmented interactively to assist probabilistic classification; cortical thickness will be determined along the entire cortical sheet as the linear distance between the outer edge of the cortical surface and the grey-white matter boundary. Ventricular size will be determined by a space filling algorithm, while shape will be characterized using LONI tools for shape decomposition and quantitative description. DWI processing routines will be developed to better account for TBI-related changes in diffusion metrics. Results from multimodal analyses will be visualized using VTK, Slicer, and other suitable platforms.
Anatomical Data
To rigorously assess workflows using the NA-MIC Kit, we will examine neuroimaging data obtained from TBI patients. MRI volumes from 202 subjects will be drawn from the LONI Image Data Archive (IDA), a comprehensive neuroimaging data archive comprised of a number of funded projects. Samples will include patients who have suffered from TBI (N=160; 22F:138M) and age-matched normal controls (N=42; 13F:29M). Mean±sd ages for males is 33.8±9.2 and for females is 33.6±9.9. T1-weighted whole brain MPRAGE volumes, T2, and, in subjects with available data sets, diffusion weighted imaging (DTI/HARDI) collected at 1.5 and 3.0T will be utilized. Additional data include a variety of MR imaging modalities and NA-MIC workflows will be crafted to accommodate them.
Expected Results
Multimodal results will be obtained using Slicer software tools, specifically developed under NA-MIC using ITK, VTK, for the analysis of neurological concomitants of TBI. Metrics will be extracted and imported into purpose built software for univariate and multivariate modeling to provide additional insights to that of previous work on the role of neuroanatomical changes occurring in TBI on outcome variables predicting degree of change and/or recovery. Several primary hypotheses using individual and repeated imaging include:
1. Cerebral atrophy (regional and global) occurs at a faster rate in diffuse vs. focal TBI;
2. Rates are dependent upon initial injury severity
3. Ongoing or progressive change continues up to 1 yr post-TBI
4. Secondary insults increase the rate and extent of the initial TBI
We will also examine age-at-lesion effects, since these factors are likely to impact on measures of the degree of loss of developmental and life-span neuroplasticity believed to follow TBI. Using DWI data, we will assess the effects of TBI on mean diffusivity, fractional anisotropy, and their potential as clinical outcome correlates. Complete multimodal data processing solutions using the NA-MIC Kit and associated tools will be made openly available, with accompanying training materials via the NA-MIC web site, and comply with the NA-MIC open-source policies.
Clinical Utility
The emphasis from the NA-MIC KIT workflows developed under this program will be on the application of the NA-MIC tools to TBI clinical practice and patient monitoring. With knowledge of general location, extent, and degree of change, such metrics can be associated with clinical measures and used to suggest viable treatment options for a subject against patterns typical of TBI patients.
Current Tasks
Considerable interactions presently exist between the NA-MIC project and LONI, resulting in numerous peer-reviewed publications. This shows the suitability of these teams to work jointly under this proposed project. Computational outcomes from this DBP will also directly contribute to other NA-MIC DBPs involving
1. mapping head and neck radiation therapy
2. tracking MRI brain changes in Huntington’s Disease
3. segmenting cardiac MR atrial wall ablations in arrhythmia patients.
UCLA investigators will interact directly with NA-MIC Core 1 and 2 PIs in developing new and refining existing processing algorithms for addressing the issues inherent to neuroimaging data from TBI patients (YR1). DBP activities at UCLA will occur in consultation with the Primary Technical NA-MIC DBP Contact and the Algorithm and Engineering Contact. Once completed and rigorously validated these workflows will
1. be applied to the TBI patient data described above (YR1-2)
2. be made available for open dissemination via the NA-MIC website (YR3)
3. form the basis for training and educational materials for NA-MIC investigators and the TBI community (YR3)
Results will be featured in presentations at scientific conferences, organized training events/workshops, etc., as a way to disseminate tool capabilities and, where possible, tutorials on how to use the NA-MIC technology for other TBI-related projects. We will attend each NA-MIC All-Hands-Meeting to discuss the DBP with NA-MIC PIs, report on developments, and progress. NA-MIC will benefit from this DBP by exposure to a unique community that often possesses multisite neuroimaging clinical trials data.
Case Analysis Goals
- Multimodal registration within and between scan sessions separated by ~6 months
- Computation of neuroanatomical measures of change per unit time in each case
- Visualization of 3D models and their degree of change
- Quantification of alterations in ventricle and WM/GM volume, surface, cortical thickness, and other morphological metrics at time 1 and time 2
- Ability to quantify changes in diffusion local to the lesions as well as overall mean FA and fiber patterns between scan sessions
- Generation of a clinical report for each TBI patient. The clinician should have the ability to store such a report and use it in conjunction with their neurological assessments to gain insight about the case. It would be nice if that report could be generated not only in printer-friendly format but also in XML (or similar format) so that it can be read without much trouble into Matlab, R, or other secondary analysis frameworks or databases.
- Semi-automatic segmentation. Certain aspects of image segmentation segmentation may need to be user-guided (e.g. to conditionalize segmentation given the presence of a lesion), though in a way that is easy for clinicians to use. The user might point and click on an area exhibiting a bleed, or some other type of hypo-/hyperintesity. A boundary contour could then be found around that area, and the segmentation model would then be weighted by the presence of that area as some unique tissue type.
- Creation of population-based atlases from multiple TBI subjects or at least reference to standardized coordinate systems
Software Requests
- The UCLA team would like to have Windows compilations of (1) the Longitudinal Lesion Comparison and (2) the Lesion Segmentation Applications Modules.
Specific Tasks In Progress
Specific tasks include the following, in reverse chronological order:
- There is some interest in a macro capability to record selections for later playback
- Full brain fiber tractography
- NIFTI 4D file read/write, external gradient information importation, and a gradient table editor for DTI
- Better interoperability between LPS/RAS file organization in Slicer vs. vTK
- Ability to set direction cosines directly in Slicer volumes module
- Timed autosave functionality of scene files and other filetypes fundamental to Slicer (saved for several time steps)
- Welcome screen should be a pop-up window in the middle of screen that could be disabled
- Integration of volume render with slice displays (rendering set aside as an option)
- Import tools for DTIStudio and TrackVis tractography files
- Creating 3D ROIs using the edge detection routine
- Use of customizable toolbars for commonly used operations/features/modules/workflows/etc
- Volumes shouldn’t be loaded differently from “other data”
- Screen shot functionality shouldn’t force a re-render
- Being able to record video sequences (at least we couldn’t readily find this capability)
- NIFTI fMRI processing capability (3D+time)
Completed
- 2010-Dec-20. ABC module has been successfully compiled for Windows and tested (use Slicer nightly build for best results). Our thanks to Marcel and to the Utah team!
Data
- Need to obtain CT and PET for each example case (Jack Van Horn, UCLA TBI team)
Results
Sample Data Sets
IMPORTANT NOTE: Use of these data for inclusion in peer-reviewed publication is provided contingent on appropriate recognition of the following UCLA investigators as study co-authors: Andrei Irimia, Micah C Chambers, Jeffry R Alger, Paul M Vespa, John D Van Horn.
- File:TBI Patient1 nrrd3.zip
- File:TBI Patient2 nrrd2.zip
- File:Patient3 DTI nrrd files.zip
- File:Patient1 UCLA 3T.zip
- File:Patient2 UCLA 3T.zip
- File:Patient3 UCLA 3T.zip
- File:Patient3 UCLA CT.zip
- File:UCLA 1 5T.zip
- File:UCLA 3T.zip
Sample Slicer Results
The following PDF contains a presentation prepared by Dr. Guido Gerig which showcases some example results on the above TBI data sets obtained using existing and newly developed Slicer processing modules.
TBI Results at UCLA
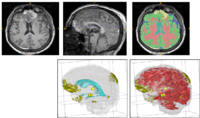
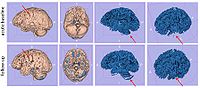
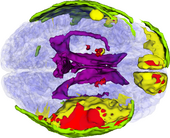
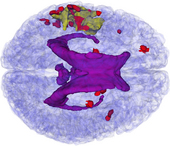
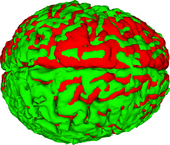
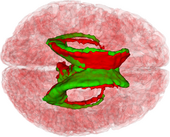
The ABC module in 3D Slicer has been applied to segment the anatomic MR volumes for Patients 1-3 in the Sample Data Sets section. In the adjacent figures, sample images of the segmentations for Patient 3 (both acute baseline and follow-up) are shown. In the first image, the WM and GM segmentations are shown for the acute baseline and follow up scans of Patient 3. We have found that, for this TBI patient, the ABC segmentation is superior to that of the EMSegmenter Module and we are excited about the ability of ABC to capture the neuroanatomical outliers of these challenging data. Many of the items of concern that remain concerning the quality of the segmentation might be addressed using suitable modifications of input parameters to ABC. For example, there seems to be some inconsistency between scans in the reconstructed gyrification of the cortex and extent of the GM/WM interface (see arrows, first and third columns). We are looking forward to learning more about the effective use of ABC to avoid these problems.
From the second figure it can be inferred that, in the case of the acute baseline scan, ABC may have failed to include the lesion in the temporal lobe as part of the WM volume. This did not occur during the segmentation of the volume acquired during the follow-up scan, where this region seems to have been more appropriately segmented (see arrows).
Longitudinal comparison of TBI using Slicer 3.6
Few non-commercial software tools are available to help clinicians analyze and solve problems of quantitative analysis and visualization. Problems on which there has been significant progress up to this point include data processing challenges, problems recognizing acute versus chronic phases of traumatic brain injury, establishing the means of performing more sensitive quantification of change, and determining extent of cortical and ventricular injury, amount of white matter injury and axonal damage and the means of associating change TBI morphometry over time with clinical outcome.
To date, the UCLA team has successfully performed co-registration of several baseline and follow-up TBI volumes, classification of healthy and injured tissues, as well as longitudinal analysis of TBI cases. Specifically, we have introduced the combined the use of multimodal TBI segmentation and longitudinal analysis using 3D Slicer. For three representative TBI cases, semi-automatic tissue classification and 3D model generation have been performed to assess longitudinal TBI evolution using multimodal volumetrics and clinical atrophy measures. Identification and quantitative assessment of extra- and intra-cortical bleeding, lesions, edema and diffuse axonal injury was also performed. 3D Slicer tools have been used to perform cross-correlation of multimodal metrics from structural imaging (structural volume, atrophy measurements, etc.) and with clinical outcome variables (time since injury, age, gender, etc.) and other potential factors predictive of recovery.
3D Slicer workflows have been found to be suitable for TBI clinical practice and patient monitoring, particularly for assessing damage extent as well as for the measurement of neuroanatomical change over time. With knowledge of general location, extent, and degree of change, such metrics computed in 3D Slicer can be associated with clinical measures and subsequently used to suggest viable treatment options for individual subjects against patterns that are typical TBI populations. Thus, the methodology that has been demonstrated up to this point using the 3D Slicer platform has the potential for significant impact upon the state of the art in TBI neuroimaging as well as upon the added benefit of TBI neuroimaging techniques from the standpoint of clinical monitoring, diagnosis and treatment.
Shown below are four images that are illustrative of this procedure for Patient 3 (MR and CT data available for download from above). A more comprehensive description of this analysis will very soon be made available in a manuscript to be submitted for peer review.
In conclusion, although the UCLA team is only 4 months into the first year of participating in the NA-MIC project, we have not only already fulfilled all specific aims for Year 1, but also made a significant amount of progress beyond these aims.
3D Slicer as a software platform for TBI
To address the urgent need for clinician-friendly TBI analysis tools, we have combined the use of multimodal, semi-automatic TBI analysis methods within 3D Slicer. To showcase the ability of the UCLA team to perform quantitative longitudinal analysis of TBI in 3D Slicer, we have analyzed three cases of semi-automatic TBI volume segmentation and 3D brain model generation while also highlighting the added clinical insight which 3D Slicer can offer.
Slicer 3 has the capability of providing visual assessment of multimodality imaging of 3D fiber tracts and morphometry in TBI. It provides the possibility for potential identification of specific targets for neurological testing enabling the clinician or researcher to deploy tests based on hypotheses derived from image analysis. Ultimately, through a more principled approach, quantitative assessments can be made.
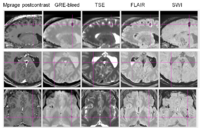
Over the past 4 months, the UCLA team has developed a sophisticated protocol for image segmentation and model generation, which has been applied to three representative TBI patients with spectacular results. This standard protocol has been confirmed to be optimal for TBI case analysis in 3D Slicer. Specifically, brain lesions adjacent to CSF were segmented from volumes acquired using FLAIR, GRE imaging, TSE T2-weighted volumes as well as DWI. Because SWI is generally superior to GRE and T2-weighted imaging to detect hemorrhagic lesions, volumes acquired using the former modality were used to identify micro-hemorrhages. Images that were additionally available in the context of our protocol were used to confirm segmentation accuracy as well as to illustrate the additional capabilities of 3D Slicer to segment images from a variety of MR data sets and combination of sequences. Throughout the past several months, the UCLA team has demonstrated the usefulness of semi-automatic segmentation tools available in 3D Slicer software, including the Atlas Based Classification (ABC) segmenter. As opposed to other specialized segmenters where access is often restricted from outside users, the ABC segmenter is freely available as a segmentation module in 3D Slicer. The method is automatic, its execution requires minimal user supervision, and its appropriateness for TBI case analysis is excellent. In addition, the ABC segmenter possesses the ability to perform co-registration of an arbitrary number of MR volumes acquired using various sequences. This makes ABC highly suitable to the UCLA multimodal TBI imaging paradigm, where as many as 12 distinct sequence types are employed in the context of a sophisticated TBI analysis protocol. At UCLA, tissue-type segmentation has already been used to calculate the total volumes of selected structure types (ventricular system, non-hemorrhagic lesions, and hemorrhagic lesions, white matter and gray matter). Volume changes have been computed as the ratio of the difference in volume between the follow-up and acute baseline time points, to the volume at the latter time point. In addition to these measures, the UCLA team has also computed five measures of atrophy, namely the bifrontal index, the bicaudate index, Evan’s index, the ventricular index and Huckman’s index. These measures as computed in Slicer have been found to be in excellent agreement with previous results available in the TBI literature.
In conclusion, over the past 4 months, we have used Slicer 3 to perform multimodal data fusion (linear co-registration, segmentation using ABC, etc.). Tissue classification with normal atlas prior, deformable (fluid) atlas to subject registration, as well as segmentation of lesions, bleedings, shunts has also been performed.
Outreach
Our outreach goals include:
- TBI Cases TBI Registration Case Library
- Develop comprehensive tutorial/how-to guide
- Design user friendly TBI workflow interface for Slicer
- Presentations/posters at OHBM, SFN, and other (inter)national meetings in year 2
- Hands-on teaching event for the DBP scientific community for year 3
- Peer reviewed publications
Outreach Events
UCLA Neurology Science Day (January 2011)
The UCLA Department of Neurology held its Third Annual Neurology Science Day on Wednesday, January 26, 2011. During the Poster Session, Andrei Irimia presented a poster to the Neurology Faculty at UCLA.
File:Irimia-UCLA-Neurology-Day-poster.pdf
TBI DBP Meeting at NA-MIC AHM (January 2011)
The UCLA and SCI teams had several fruitful and friendly meetings during the All Hands Meeting in Salt Lake City. The first of these meetings was attended by about 30 researchers, including Guido Gerig, Marcel Prastawa, Bo Wang, Sylvain Gouttard, Andrei Irimia, Micah Chambers, Stephen Aylward, Hans Johnson, and their teams. This part of the meeting focused on how Slicer tools can be used to study pathology, with a focus on Huntington Disease and TBI. In the opening of the meeting, Guido raised the question of what Slicer tools are already available to assess longitudinal changes. Hans Johnson expressed his interest in Slicer and commented on the possible activity of using the platform for the development of clinical trials.
For the TBI project, the Utah team is interested in obtaining radiological TBI case descriptions from UCLA radiologists and in having more input from UCLA as to how various types of TBI images (e.g. Flair, GRE bleed, etc.) should be interpreted. Such radiological case descriptions would allow one to better interpret TBI data and to determine the class of pathology to which various features belong, based on a comparative examination of different scans. Another request from the Utah team is to have patient histories for the subjects that we provide for them. They would like to use this information in conjunction with the case descriptions to learn more about the type of lesions that they are studying, and to obtain a sense of how the patient history and case evolution is reflected in the anatomical changes that are apparent from the scans.
One concern that was expressed regards the ability of this project's time frame to accommodate both algorithm development as well as the development of clinically oriented tools and measures which require the output of the former from the very beginning. For example, we need to develop longitudinal analysis tools but the algorithms to do TBI segmentation are a prerequisite to this, although it may take years to develop the latter. Andrei pointed out to the possibility of using mild TBI cases in the beginning in order to develop longitudinal analysis tools, while the Utah team is developing more complex and robust methods that can handle difficult cases.
Five goals of clinical interest were discussed during the meetings, namely (1) the ability to do TBI tissue classification, (2) the ability to perform volumetric analysis to allow for neuroanatomic assessment, (3) the ability to track the fate of lesions and pathology using automatic segmentation, (4) the ability to generate quantitative output in the form of a report to demonstrate that the use of imaging correlates with improvement in outcome scores, and (5) the ability to easily quantify WM tract distortion and change, as well as how this distortion evolves over time.
One item that was pointed out to the entire audience is that the focus of Slicer is on subject-level rather than population-level analysis, in spite of existing clinical interest in performing statistical studies of population groups. However, one item which clinicians would find appealing is precisely the ability to compute anatomical measures at the subject level, particularly because these subject-level measures can then be used for population-level statistical analysis outside Slicer. Although such population-level analyses may not be within the scope of Slicer, it may be useful to take their specifications into account in the context of formulating goals and strategies for the collaboration.
ABC segmentations of TBI volumes were reviewed in a separate session at the meeting, and a number of plans for the future were formulated. One item that was discussed was the selective use of image channels to perform TBI segmentation. Depending on the type of pathology that is the subject of focus, it may be of interest to include in the segmentation process only those scans in which the pathology is most obvious. The two teams decided to collaborate on how best to accomplish this.
A number of technicalities related to ABC and to segmentation in general were discussed, with the end conclusion being that the results thus far generated with ABC can probably be much improved upon after some exploratory analysis. Team members agreed to keep in touch by email, phone, teleconference, etc. It was agreed upon that there should be some type of periodical face-to-face interaction between the teams at UCLA and SLC, possibly every several months.
TBI DBP Conference Call with Utah Team (September 9, 2011)
The UCLA and Utah teams had a productive conference call on September 9, 2011. All progress up to this date was reviewed, including (1) integration of MRI Watcher into Slicer, (2) the upcoming version of Slicer in QT, (3) the conference paper that was submitted to SPIE, and (4) the availability of CT data for TBI patients. The two teams also reviewed various technical items, including (5) the calculation and display of Jacobian deformation maps for TBI between acute and chronic time points for use by clinicians, (6) the issue of how to address changes in topology between time points, (7), possible relaxation of diffeomorphic constraints for the purpose of quantifying the deformation field, (8), the step-by-step TBI tutorial that was presented by Andrei Irimia at the Boston June Meeting, (9) possible future coordination between the Utah and UNC/Kitware/GA Tech teams. The overall conclusion from the meeting was that much progress has been achieved this year and that we are looking forward to presenting our accomplishments in Salt Lake City at the Annual All-Hands Meeting.
NA-MIC Conference Call (October 27, 2011)
During the NA-MIC conference call with Dr. Kikinis and his team at BWH, Dr. Van Horn highlighted our excellent research progress throughout the past year, including publication of a paper in the Journal of Neurotrauma that uses Slicer, one publication accepted for MICCAI 2012 based on a study directed by Guido Gerig at the University of Utah, two manuscripts on TBI geometric metamorphosis based on research directed by Stephen Aylward from Kitware, one paper based on research directed by Yifei Lou and Allen Tannenbaum on deformable registration, and other conference abstracts and posters.
TBI DBP Conference Call (November 10, 2011)
A conference call was held with the Kitware, GA Tech and Utah teams regarding activities planned for the NA-MIC All Hands Meeting in Salt Lake City. While there, the three teams will collaborate on atlas based classification, nonrigid registration algorithms, and on several manuscripts in progress.
NA-MIC Conference Call (November 16, 2011)
Dr. Van Horn had a conference call with Tina Kapur regarding the upcoming meeting in January. Our recent progress was highlighted, including the publication of a paper in the Journal of Neurotrauma, conference abstracts and proceedings, posters and other activities.
Investigators
UCLA
* John Darrell Van Horn, M.Eng., Ph.D. - Principal Investigator of the DBP * Andrei Irimia, Ph.D. (DBP engineer, postdoctoral scholar) [2] * Micah Chambers, M.S. (graduate student) * David Hovda, Ph.D. (investigator) * Paul Vespa, M.D., F.C.C.M., F.A.A.N. (investigator) * Arthur W. Toga, Ph.D. (investigator) * Jeffry Alger, Ph.D. (investigator)
University of Utah
* Guido Gerig, Ph.D. (lead technical contact) * Marcel Pastrawa, Ph.D. (research scientist) * Sylvain Gouttard, M.S. (postdoctoral scholar) * Bo Wang, B.S. (graduate student)
Kitware/UNC Chapel Hill
* Stephen Aylward, Ph.D. (director of medical imaging) * Danielle Pace * Ilknur Kabul * Martin Styner, Ph.D. (lead algorithm and engineering contact)
Georgia Tech
* Allen Tannebaum, Ph.D. (Principal Investigator of the Algorithm Core) * Yifei Lou, Ph.D. (postdoctoral scholar)
Harvard Medical School
* Ron Kikinis, M.D. (Principal Investigator) * Sonja Pujol, Ph.D.
Awards and Honors
- Andrei Irimia obtained the First Prize in the Summer Tutorial Contest of the National Alliance for Medical Image Computing [3]
- Andrei Irimia's visualization of TBI using Slicer has been selected to be featured on the cover of the Journal of Neurotrauma for a special issue on bioengineering for TBI
- Andrei Irimia was awarded the first prize in the Fine Science Award Competition by the Brain Research Institute at UCLA for excellence in postdoctoral neuroscience research
- Andrei Irimia obtained a competitive travel award to present TBI research at a Keystone Symposium on the Molecular and Clinical Biology of TBI to be held in Keystone, Colorado in February 2012
- Andrei Irimia obtained a travel scholarship to present a TBI-related poster at the Dynamic Days 2012 conference to be held in Baltimore, MD in January 2012
Publications
Journal Articles
- Andrei Irimia, Micah C. Chambers, Jeffry R. Alger, Maria Filippou, Marcel W. Prastawa, Bo Wang, David A. Hovda, Guido Gerig, Arthur W. Toga, Ron Kikinis, Paul M. Vespa, John D. van Horn (2011) Comparison of acute and chronic traumatic brain injury using semi-automatic multimodal segmentation of MR volumes. Journal of Neurotrauma [4]
Proceedings
- Bo Wang, Marcel W. Prastawa, Suyash P. Awate, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. Van Horn and Guido Gerig (2012) Segmentation of serial MRI of TBI patients using personalized atlas construction and topological change estimation Proceedings of the Tenth International Symposium on Biomedical Imaging (ISBI 2012), May 2-5, 2012, Barcelona, Catalonia, Spain
- Bo Wang, Marcel W. Prastawa, Andrei Irimia, Micah C. Chambers, Paul M. Vespa, John D. van Horn, Guido Gerig (2012) A patient-tailored segmentation framework for longitudinal MR images of traumatic brain injury Proceedings of the Twenty-Fifth International Conference of SPIE—The International Society for Optical Engineering, February 4-9, 2012, San Diego, California, USA
- Marc Niethammer, Gabriel L. Hart, Danielle F. Pace, Micah C. Chambers, Andrei Irimia, John D. van Horn, Stephen R. Aylward (2011) Geometric metamorphosis Proceedings of the Fourteenth International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2011), September 18-22, 2011, Toronto, Ontario, Canada
Abstracts
- Andrei Irimia, Micah C. Chambers, Paul M. Vespa, Arthur W. Toga, John D. van Horn (2011) Cortical network visualization and analysis in traumatic brain injury using multimodal neuroimaging Proceedings of the Eighteenth Joint Symposium on Neural Computation, June 4, 2011, Institute of Neural Computation, University of California, San Diego, La Jolla, California, USA
- Andrei Irimia, Micah C. Chambers, Maria Filippou, Jeffry R. Alger, Marcel W. Prastawa, Bo Wang, Silvain Gouttard, Sonia M. A. Pujol, Stephen R. Aylward, David A. Hovda, Guido Gerig, Arthur W. Toga, Ron Kikinis, Paul M. Vespa, John D. van Horn (2011) Three‐dimensional calculation and quantification of morphometric and volumetric cortical atrophy indices of widespread clinical use from MRI volumes of traumatic brain injury using 3D Slicer Proceedings of the Forty‐First Annual Meeting of the Society for Neuroscience (SfN 2011), November 12-16, 2011, Washington, District of Columbia, USA
- Andrei Irimia, John D. van Horn, Micah C. Chambers, Marcel W. Prastawa, Silvain Gouttard, Paul M. Vespa, David A. Hovda, Jeffry R. Algers, Sonia M. A. Pujol, Guido Gerig, Stephen R. Aylward, Arthur W. Toga, Ron Kikinis (2011) Automatic multimodal MR image segmentation for the clinical assessment of traumatic brain injury in 3D Slicer Proceedings of the Seventeenth Annual Meeting of the Organization on Human Brain Mapping (OHBM 2011), June 26-30, 2011, Quebec City, Canada
Presentations
- Andrei Irimia, Micah C. Chambers, John D. van Horn (2011) Automatic segmentation of traumatic brain injury MRI volumes using atlas based classification and 3D Slicer Tutorial Presentation Session of the Seventh Annual Project Week of the National Alliance for Medical Image Computing, June 20-24, 2011, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA
- Andrei Irimia, Micah C. Chambers, John D. van Horn (2011) Analysis and visualization of traumatic brain injury using 3D Slicer within the NA-MIC Collaboration Invited Presentation at the Seventh Annual Project Week of the National Alliance for Medical Image Computing, June 20-24, 2011, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA
- Andrei Irimia, Micah C. Chambers, John D. van Horn (2011) Traumatic brain injury processing, analysis and visualization. Laboratory of Neuro Imaging, University of California, Los Angeles, June 3, 2011, Los Angeles, California, USA
- Andrei Irimia, John D. van Horn, Micah C. Chambers (2011) Longitudinal analysis of traumatic brain injury using 3D Slicer. Brain Injury Research Center, University of California, Los Angeles, March 25, 2011, Los Angeles, California, USA
- Andrei Irimia, John D. van Horn, Micah C. Chambers, Marcel W. Prastawa, Bo Wang, Silvain Gouttard, Paul M. Vespa, David A. Hovda, Jeffry R. Alger, Sonia M. A. Pujol, Guido Gerig, Stephen R. Aylward, Arthur W. Toga, Ron Kikinis (2011) Clinically-driven multimodal imaging of traumatic brain injury using semi-automatic segmentation in 3D Slicer. Third Annual Neurology Science Day, University of California, Los Angeles, January 26, 2011, Los Angeles, California, USA
- John D. van Horn, Andrei Irimia, Micah C. Chambers, Arthur W. Toga, Jeffry R. Alger, Paul M. Vespa, David A. Hovda (2011) UCLA Traumatic Brain Injury. Seventh All Hands Meeting of the National Alliance for Medical Image Computing (NA-MIC AHM), January 10-14 2011, Salt Lake City, Utah, USA
Links
- UCLA Laboratory of Neuro Imaging(LONI)
- Utah Center for NeuroImage Analysis
- Traumatic Brain Injury Portal
- TBI IMPACT Mission