Difference between revisions of "DBP3:Utah:AutoWallSeg"
| Line 134: | Line 134: | ||
* CARMA manual segmentations exclude portions of the anatomy such as pulmonary veins, mitral valve, and the LA appendage, while the proposed algorithm makes no distinction between these structures and myocardium. | * CARMA manual segmentations exclude portions of the anatomy such as pulmonary veins, mitral valve, and the LA appendage, while the proposed algorithm makes no distinction between these structures and myocardium. | ||
* The Slicer implementation of the proposed algorithm does not allow for the inner level-set surface to move away from the endocardial segmentation surface. Thus, there can be no refinement of the boundary between blood-pool and endocardium in the LA wall segmentation. | * The Slicer implementation of the proposed algorithm does not allow for the inner level-set surface to move away from the endocardial segmentation surface. Thus, there can be no refinement of the boundary between blood-pool and endocardium in the LA wall segmentation. | ||
| − | * The proposed algorithm is subject to "leakage" of the LA wall boundary into surrounding structures. This is presumably because the proposed method models endocardium as an intensity distribution, which we know is not well-separated from intensity distributions in the surrounding tissues. Artifact, partial- | + | * The proposed algorithm is subject to "leakage" of the LA wall boundary into surrounding structures. This is presumably because the proposed method models endocardium as an intensity distribution (?), which we know is not well-separated from intensity distributions in the surrounding tissues. Artifact, partial-volume effects, MRI coil sensitivity inhomogeneities, and enhancement of surrounding anatomy all contribute to this problem. |
| + | * The time to run the proposed automated algorithm is almost 50% of the time necessary for average manual segmentation. | ||
| − | Automatic segmentation of the LA wall from LGE MRI is an ill-posed and very difficult problem. | + | '''In Summary:''' Automatic segmentation of the LA wall from LGE MRI is an ill-posed and very difficult problem. The bottom-line is that the LA boundary is just very difficult to see in these images. Finding reliable estimates of the LA wall boundary requires significant experience in reading cardiac LGE-MRI and a good working knowledge of cardiac anatomy. Intensity models that separate normal myocardium from enhanced myocardium and surrounding tissues have all proven difficult to estimate reliably from LGE MRI. In conclusion, for LGE-MRI at its current resolution and SNR, we do not expect to find a completely automated algorithm for LA wall. Instead, we will focus our efforts on automation of the endocardial segmentation, where we can rely on shape and intensity information from the blood pool, which is more easily modeled in LGE MRI and MRA. |
Revision as of 18:45, 3 November 2011
Home < DBP3:Utah:AutoWallSegContents
Automatic Segmentation of LA Wall from Endocardial Segmentation
Automatic Wall Segmentation from GA Tech
GA Tech produced a slicer module to automatically segment the left atrial wall, given the original data AND the endocardial(blood pool) segmentation. Below is some evaluation of those segmentations.
Visual
(click on thumbnails to view full size)
Slice
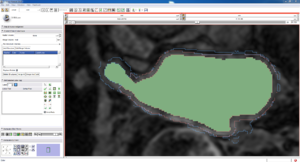
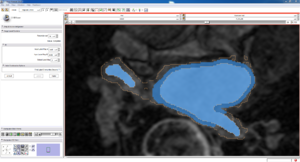
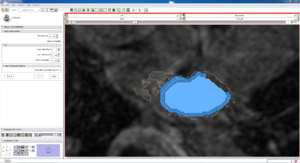
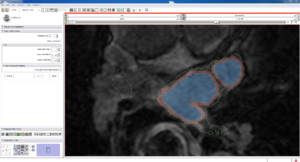
| Auto, Endo, and Manaul Wall (01 3mo s5) | Auto, Endo, and Manual Wall (01 3mo s4) | Auto, Endo, and Manual Wall (01 3mo s1) | Auto, Endo, and 4 Pixel Dilation from Endo (01 3mo s1) |
|---|---|---|---|
| This image shows how the automatic wall segmentation creates a complete surface around the endo - while a manual segmentor would cut off the wall around the veins (although the location this cutoff decision is arbitrary). | This image shows where the automatic segmentation surrounds a vein, while the manual segmentor decided to not surround the vein. | This image shows some island artifacts that may be the result of the process being done in 3D (leaking from the slice above) - note that the data below the islands is lighter. | Here we show a 4 pixel dilation from the endo compared to the automatic segmentation. This also image shows more islands. |
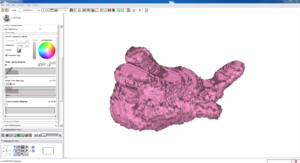
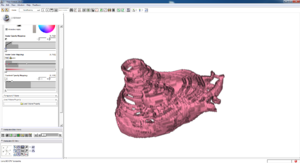
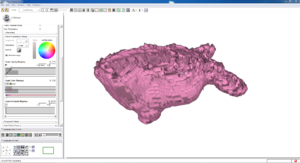
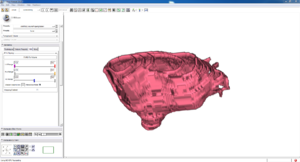
Isosurface
Note how the automatic segmentation creates a smooth/complete (mostly) surface on the top side of the atrium, while the bottom has gaps similar to the manual.
| Auto Wall Segmenation Isosurface (01 3mo s5) | Manual Wall Segmentation Isosurface (01 3mo s5) |
|---|---|
Comparison Statistics
Inter Auto Seg Comparison
Before getting the automatic wall segmentation code we had done a small study on our manual segmentations (see LA Segmentation reproducibility - intra and inter observer), this involved 64 total Endo + Wall Segmentations (4 different segmentors segmenting the same scan, for 16 scans).
To compare them we used the STAPLE algorithm to create a "ground truth" and then calculated overlap and accuracy, among other things, according to that ground truth.
We did the same analysis, but on the automatic wall segmentations (each derived from the different manual endo segmentations), and compared it to the original manual segmentation data.
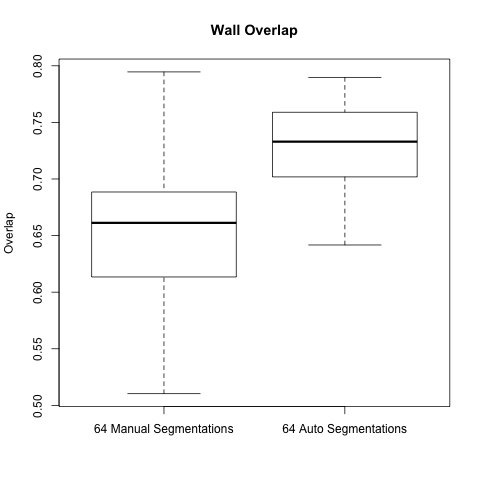
Below are boxplots of the two data sets, for both overlap and accuracy.
For two sets S1 and S2, overlap is computed: (2 * ||S1 ^ S2|| ) / ( ||S1|| + ||S2|| )
For two sets S1 and S2, accuracy is computed: (||S1 ^ S2|| + ||!S1 ^ !S2|| ) / (||S1|| + ||S2||)
(For brevity: ^ is intersection, U is union, and !S is the complement of S)
As the graphs show the automatic wall segmentations have much better overlap and accuracy with their STAPLE produced ground truths.
However the most these results seem to say is that the automatic algorithm is more consistent than manual segmentations. We hope to run a new comparison to be able to really evaluate the performance of the automatic segmentation.
A side note is that the manual segmentations are actually very consistent when compared to the automatic segmentations - one would expect the gap to be larger.
Original data: File:Autoseg.xlsx, File:Manual Compare Results.xlsx
Auto to Manual Overlap Comparison
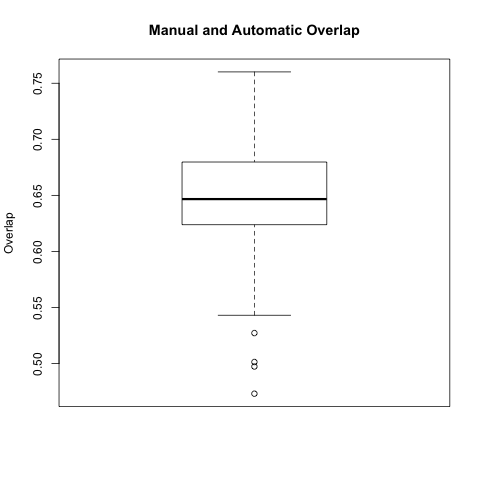
In this comparison we took each automatic segmentation produced using a manual endo segmentation and compared it with the corresponding manual wall segmentation.
The box plot below is for all 64 automatic-manual segmentation pairs.
Original data: File:Autoseg pairs.xlsx
Comparison with Epi Wall including Pulmonary Veins
The preceding analysis was done with the final product wall from a manual segmentation session, which has all of the pulmonary veins removed. Pulmonary vein removal, however, is somewhat arbitrary and the GA Tech results don't incorporate vein removal in the process. To perform a more fair comparison, we did a second analysis using the wall segmentation with veins still attached (in our manual data we subtracted the endocardial segmentation from the epicardial segmentation).
Visual
| good slice | bad slice | |
|---|---|---|
| Best overlap | ||
| Worst overlap |
Statistical
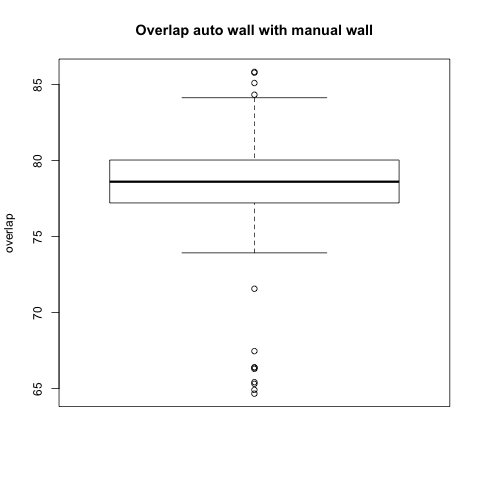
|
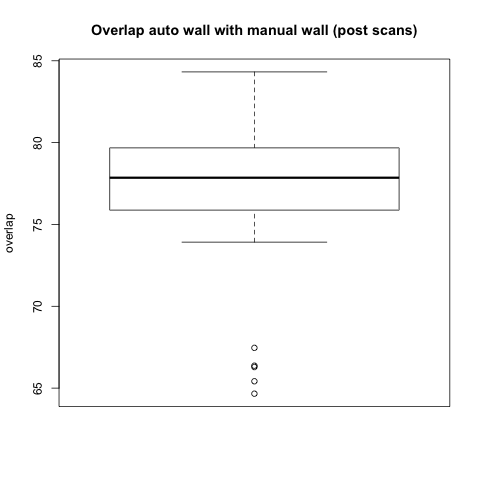
|
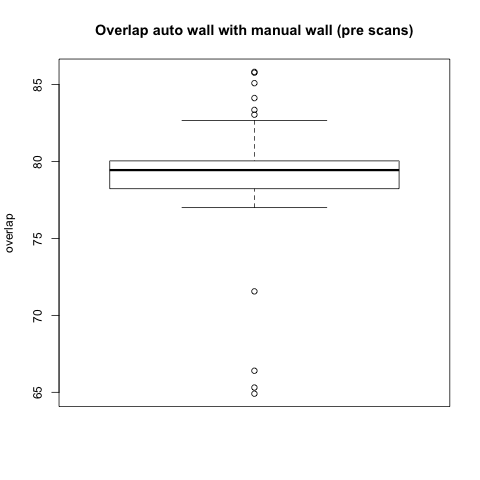
|
Timing
In release mode each automatic wall segmentation takes about 3-4 minutes. By comparison, a manual wall segmentation takes about 8 minutes.
A more detailed analysis gave the following timing results over our 73 patient cohort:
- mean duration: 223.98 sec (3 m 43 s)
- standard deviation: 46.43 sec
Note: This is time spent in user space, time spent in kernel space was ignored (usually less than a second).
Conclusions and Discussion
Our main conclusion from the preceding analysis are that segmentations produced by the proposed LA wall segmentation algorithm do not show good agreement with LA wall segmentations that are produced manually at CARMA. Important observations on manual vs. the proposed algorithm are listed below.
- CARMA manual segmentations exclude portions of the anatomy such as pulmonary veins, mitral valve, and the LA appendage, while the proposed algorithm makes no distinction between these structures and myocardium.
- The Slicer implementation of the proposed algorithm does not allow for the inner level-set surface to move away from the endocardial segmentation surface. Thus, there can be no refinement of the boundary between blood-pool and endocardium in the LA wall segmentation.
- The proposed algorithm is subject to "leakage" of the LA wall boundary into surrounding structures. This is presumably because the proposed method models endocardium as an intensity distribution (?), which we know is not well-separated from intensity distributions in the surrounding tissues. Artifact, partial-volume effects, MRI coil sensitivity inhomogeneities, and enhancement of surrounding anatomy all contribute to this problem.
- The time to run the proposed automated algorithm is almost 50% of the time necessary for average manual segmentation.
In Summary: Automatic segmentation of the LA wall from LGE MRI is an ill-posed and very difficult problem. The bottom-line is that the LA boundary is just very difficult to see in these images. Finding reliable estimates of the LA wall boundary requires significant experience in reading cardiac LGE-MRI and a good working knowledge of cardiac anatomy. Intensity models that separate normal myocardium from enhanced myocardium and surrounding tissues have all proven difficult to estimate reliably from LGE MRI. In conclusion, for LGE-MRI at its current resolution and SNR, we do not expect to find a completely automated algorithm for LA wall. Instead, we will focus our efforts on automation of the endocardial segmentation, where we can rely on shape and intensity information from the blood pool, which is more easily modeled in LGE MRI and MRA.