DBP3:Utah:RegSegPipeline
From NAMIC Wiki
Home < DBP3:Utah:RegSegPipeline
back to DBP3 home
Contents
The CARMA DBP: MRI-based study and treatment of atrial fibrillation
Pilot Studies on a Registration & Segmentation Pipeline & Workflow
Alex Zaitsev, Dominik Meier, Ron Kikinis
Main processing pipeline
To facilitate the workflow, we can place all the automated steps at the beginning and cluster interactive elements at the end. Exception is the cropping step required as input for nonrigid registration. 1. N4 bias field correction for the MRI (surface coils):
- Input: MRI_pre and MRI_post, each run separately with the same parameters below; Output: MRI_pre_n4 and MRI_post_n4
- Module used: N4 ITK; Parameters: convergence: 1e-5, iterations: 50,40,30,20, shrink factor: 3
- Comments: a run on entire image gives some benefit that may be improved with masking: again the dominant intensity dropoff from the surface coil occurs along the chest wall and ribcage. Even if that is not the structure of interest, it is the low-freq. variation the bias correction algorithm is searching for, and masking that out can be counter-productive: via masking we may end up with a smoother image, but the intensity variations removed were not caused by the coil but are actually true signal.
2. registration MRA ->MRI (both pre and post)
- Input: MRI_pre_n4 (fixed) and MRA_pre ; Output: Xf1_pre_MRA-MRI.tfm"
- Input: MRI_post_n4 (fixed) and MRA_post ; Output: Xf2_post_MRA-MRI.tfm"
- Module used: BRAINSfit
- Module 'Parameters:' convergence: 1e-5, iterations:
- Comments: from the example dataset (P1) we infer that the two scans tend to have little initial alignment, hence initialization steps are not recommended. We choose registration parameters such that the chance of the registration making the alignment worse is minimized. Decide in a subsequent review/QC step if we keep the transform or the original pose. The MRA contains the same FOV and has surrounding structures (liver, chest, spine etc) visible also, despite lower intensities. A global affine is thus not necessarily going to benefit from masking the heart, unless the relative motion of the heart becomes the dominant reason for misalignment. We tried masking with both BrainsFit and RobustMultires modules. Both failed to provide better alignment with masking.
3. registration follow-up -> baseline: phase 1: AFFINE
- most reliably done on the post contrast MRI.
- DOF up to 12, because image is captured at different phases in the breathing/cardiac cycle
- Module used: BRAINSfit
- fixed image: LGEMRI_pre, moving image: LGEMRI_post, initialize: Moments align, DOF: rigid + scaleVersor
- defaults, except: sample points:200,000;
- done on the entire image. The surrounding structures are useful in constraining the solution transform and should provide a more robust behavior. Cropping down to the cardiac only ROI is deferred to the nonrigid registration in phase 2.
- Note: the IS FOV can differ, e.g. how much of the liver is included. If the two exams differ significantly (>30%) in that content, the above affine could fail and a prior cropping step is then suggested to better match image content before registration. Resampling to isotropic voxel size at this stage is also advantageous but will generate very large files > 100MB due to the full FOV.
- registration follow-up -> baseline: phase 2: nonrigid / BSPLINE
- crop to volume of interest: CropVolume module
- use Resample to isotropic voxelsize option.
- register cropped volumes follow-up -> baseline, using above Affine as starting point (initialization)
- Module: BRAINSfit
- fixed image: LGEMRI_pre, moving image: LGEMRI_post, initialize: Moments align, DOF: rigid + scaleVersor
- ROI definition (manual box ROI or automated via atlas)
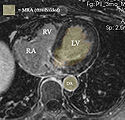
- segmentation of LA from MRA -> inner wall
- as a dynamic image the MRA contains significant spread and likely requires interactive segmentation/thresholding to yield a satisfactory LA volume
- Module used: Editor: thresholding or thresholding within Volumes thresholding option within Display tab, use iron colormap & low alpha setting to check for ventricular wall borders.
- cropping and island removal
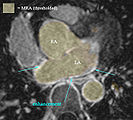
- LA wall segmentation
- very small structure, most reliably done manually direct. Starting with automation may yield more effort on post-edits
- Module used: Editor: manual outline
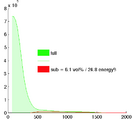
- segmentation of enhancement within LA wall: intensity statistics. An atlas-based set of intensity distributions may be more meaningful here than a simple Otsu, because both amount and location of enhancement is unknown and can in theory be 0.
Example Cases
- Example Case P2 : pre-post registration
- This example contains significant MRI (motion?) artifacts that require dedicated processing to isolate the structures of interest
- E.g. FOV includes much more of liver on pre exam, which needs to be cropped to amount matching the follow-up