Home < Downloads
The following is a collection of electronic resources provided by NA-MIC. This includes software, data, tutorials, presentations, and additional documentation.
Software

|
Download Slicer
A general purpose biomedical computing application with extensive built-in visualization and analysis capabilities, accessible through an easy to use graphical interface.
|
|
Download the NA-MIC Kit, including Slicer
The NA-MIC Kit is a free open source software platform. The NA-MIC Kit is distributed under a BSD-style license without restrictions or "give-back" requirements and is intended for research, but there are no restrictions on other uses. It consists of the 3D Slicer application software, a number of tools and toolkits such as VTK and ITK, and a software engineering methodology that enables multiplatform implementations.
|
Data

|
Brain: Multi-modality (sMRI, DTI, fMRI) from Schizophrenia Study
There are 20 cases: ten are Normal Controls and ten are Schizophrenic. Each case includes a weighted T1 scan, a weighted T2 scan, an fMRI scan, a DTI volume, the DWI with 51 directions, and several masks and labelmaps. Available from Harvard.
|

|
Brain: 2-4 Year Old from Autism Study
Data for 2 autistic children and 2 normal controls (male, female) scanned at 2 years with follow up at 4 years from a 1.5T Siemens scanner. Files include structural data, tissue segmentation label map and subcortical structures segmentation. Available from UNC.
|
|
Brain: White Matter Lesions for Lupus Study
Data for 5 cases of Lupus White Matter Lesion patients. The data is co-registered. Each case contains: T1-weighted, T2-weighted, FLAIR, and masks for brain and lesions. Available from MIND.
|

|
Prostate: 5 robot-assisted intervention cases for Prostate Cancer
MRI Prostate data. 5 datasets, with pre-operative and intra-operative scans (biopsy and seed placement procedure). Acquired at National Institute of Health (Principal Investigators: Camphausen, Kaushal and Pinto).
|

|
Prostate: 10 cases
MRI Prostate data. 10 datasets, including a derived segmentation series with labelmaps. Available from Harvard.
|
|
Prostate: Transrectal Tutorial Dataset
Transrectal Prostate Biopsy Tutorial Dataset. Walks the user through: Calibration (calibration image for the APT-MRI device), Segmentation (prostate MRI image and seeds for random walk segmentation algorithm), Targeting (target planning prostate MRI image), and Verification (needle insertion verification image). Available from Queens.
|

|
Spine Phantom: PerkStation Tutorial Dataset
Perkstation Tutorial Dataset. Available from Queens.
|

|
Visible Human Datasets
Visible Human Datasets with some post-processing. Available from Iowa.
|

|
Registration Case Library Home Page
New and growing (Oct. 2009 - Sept. 2011) list of image datasets for testing 3DSlicer registration methods & modules. Images range from brain to abdominal to musculoskeletal, modalities range from MRI, CT to PET. Data includes raw image data (NRRD), registration task description & discussion, results, parameter preset files and step-by step tutorials. Built for the clinician researcher to find a related image registration problem and thus provide a starting point for registration parameters and strategies.
|
Tutorials
Tutorials for Biomedical Engineers and Clinical Research Users of the NA-MIC Kit (PDF and PPT downloads)
|
Stochastic Tractography to extract, visualize and quantify white matter connections from Diffusion Images in Schizophrenia Study
The python stochastic tractography module contains the tools necessary to extract, visualize and quantify white matter connections from DTI images. It seeds nerve fiber bundles from regions of interest (ROIs) based on DWI images. Unlike streamline tractography, stochastic tractography uses a probabilistic framework to perform tractography.
|

|
Classification of White Matter Lesions for Lupus
This tutorial demonstrates an automated, multi-level method to segment white matter brain lesions in lupus. Following this tutorial, you’ll be able to load scans into Slicer3, and segment and measure the volume of white matter lesions on the provided data-set.
|

|
Trans-rectal MR guided prostate biopsy and PerkStationSlicerTutorial
This tutorial will teach you how to perform MR-guided prostate biopsy using MR-compatible trans-rectal robot with SLICER.
|

|
ARCTIC: Automatic Regional Cortical ThICkness Analysis for Autism
Following this tutorial, you will be able to perform an individual analysis of regional cortical thickness. You will learn how to load input volumes, run the end-to-end module ARCTIC to generate cortical thickness information and display output volumes.
|
|
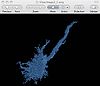
Confocal Microscopy
Guiding you step by step through the process of loading confocal microscopy data, working with that data, and creating a 3D model for visualization.
|
|
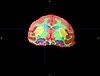
Non-human Primates Segmentation Tutorial
The objective of this tutorial is to demonstrate how to use EM Segmenter to segment non-human primate images.
|
|
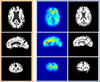
Hammer Registration for Brain MRI
Presents HAMMER registration algorithm and introduces how to use HAMMER in Slicer3.
|

|
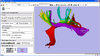
Centerline Extraction of Coronary Arteries using VMTK
Guiding you step by step through the process of centerline extraction of Coronary Arteries in a cardiac blood-pool MRI using VMTK based Tools.
|
|
EM Fiber Clustering
This module clusters a set of input trajectories into a number of bundles, generates arc length parameterization by establishing the point correspondences and reports diffusion parameters along the bundles as well as the membership probability of each trajectory in each cluster. The module requires specification of seed trajectories (or initial centerlines) as representatives of the desired bundles.
|
Additional Tutorials



















