Difference between revisions of "EMSegment"
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (7 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery> | <gallery> | ||
| − | Image:PW2009-v3.png|[[2009_Summer_Project_Week| | + | Image:PW2009-v3.png|[[2009_Summer_Project_Week#Projects|Projects List]] |
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * Sylvain Jaume | + | * Sylvain Jaume, Polina Golland (MIT Computer Science and Artificial Intelligence Laboratory) |
* Nicolas Rannou, Steve Pieper, Ron Kikinis (Harvard Medical School, Brigham and Women's Hospital) | * Nicolas Rannou, Steve Pieper, Ron Kikinis (Harvard Medical School, Brigham and Women's Hospital) | ||
| + | * Koen Van Leemput (Harvard Medical School, Massachusetts General Hospital) | ||
| + | ==Collaborators== | ||
| + | * Andriy Fedorov (Harvard Medical School, Brigham and Women's Hospital) | ||
| + | * Vidya Rajagopalan, Chris Wyatt (Virginia Tech, Wake Forest University) | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 15: | Line 19: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
The goal of the EMSegment project is to create a segmentation module in Slicer3 that offers a high productivity and reliability to the clinician. | The goal of the EMSegment project is to create a segmentation module in Slicer3 that offers a high productivity and reliability to the clinician. | ||
| − | The | + | The targeted application is the segmentation of MRI images of the brain. The EMSegment module allows the segmentation of other anatomical |
regions as long as a statistical atlas is available. | regions as long as a statistical atlas is available. | ||
| Line 31: | Line 35: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | We developed 3 new features to improve the visualization of parameters in the EMSegment | |
| − | + | and to help the user select the optimum values: | |
| − | + | ||
| − | + | * Intensity Normalization | |
| + | * Intensity Distribution by 2D Gaussian | ||
| + | * Global Priors | ||
| + | |||
| + | Our development has been committed into the Slicer3 trunk and will be available in Slicer 3.5. | ||
| + | As a side project, we developed a module for MRI Bias Field Correction: [https://www.slicer.org/wiki/Modules:MRIBiasFieldCorrection-Documentation-3.5 MRIBiasFieldCorrection] | ||
| + | We provided support to the NA-MIC group studying alcohol and stress interaction in primates, | ||
| + | and we have tested the module on monkey datasets. | ||
| + | Besides, a Command Line version of this module has been developed to enable automated Testing in Nightly builds. | ||
</div> | </div> | ||
</div> | </div> | ||
<div style="width: 97%; float: left; padding-right: 3%;"> | <div style="width: 97%; float: left; padding-right: 3%;"> | ||
| + | |||
| + | ==New Features== | ||
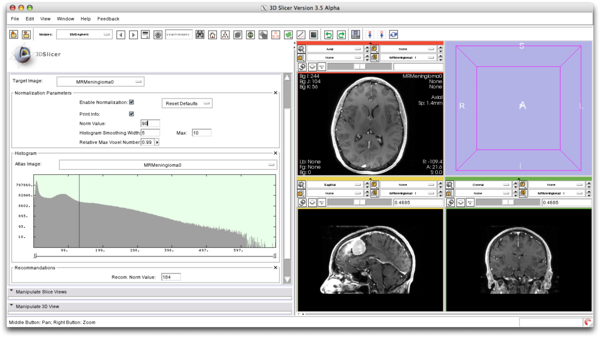
| + | ===Intensity Normalization=== | ||
| + | We compute the normalization parameter for the atlas based on the value selected by the user. | ||
| + | |||
| + | [[File:EMSegment_Intensity_Normalization.png|600px]] | ||
| + | |||
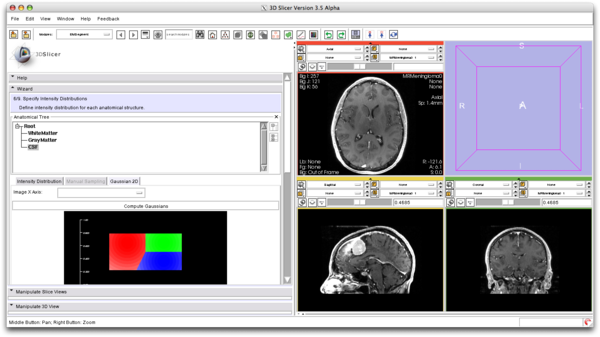
| + | ===Intensity Distribution Visualization=== | ||
| + | We model the image intensities using a Gaussian distribution for every class. | ||
| + | |||
| + | [[File:EMSegment_Gaussian_2D.png|600px]] | ||
| + | |||
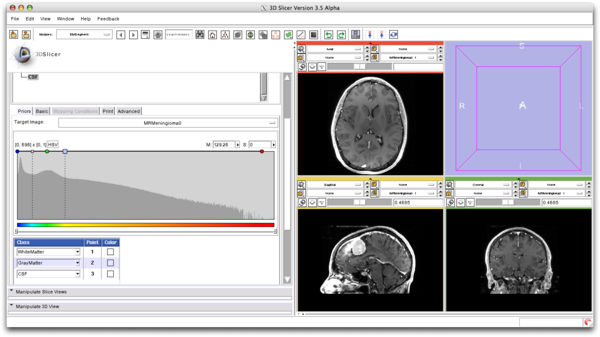
| + | ===Global Prior Visualization=== | ||
| + | We compute the Global Priors based on the selection in the intensity histogram. | ||
| + | |||
| + | [[File:EMSegment_Global_Prior_Visualization.png|600px]] | ||
==Collaboration== | ==Collaboration== | ||
Latest revision as of 17:41, 10 July 2017
Home < EMSegmentKey Investigators
- Sylvain Jaume, Polina Golland (MIT Computer Science and Artificial Intelligence Laboratory)
- Nicolas Rannou, Steve Pieper, Ron Kikinis (Harvard Medical School, Brigham and Women's Hospital)
- Koen Van Leemput (Harvard Medical School, Massachusetts General Hospital)
Collaborators
- Andriy Fedorov (Harvard Medical School, Brigham and Women's Hospital)
- Vidya Rajagopalan, Chris Wyatt (Virginia Tech, Wake Forest University)
Objective
The goal of the EMSegment project is to create a segmentation module in Slicer3 that offers a high productivity and reliability to the clinician. The targeted application is the segmentation of MRI images of the brain. The EMSegment module allows the segmentation of other anatomical regions as long as a statistical atlas is available.
Approach, Plan
Our algorithm builds upon the Expectation Maximization theory and is structured to let the clinician make the most efficient use of his/her anatomical knowledge. Because of our intuitive visualization of probabilities, the user can understand the probabilistic features of his/her data and can efficiently tune the parameters to obtain the most accurate segmentation. To meet the time constraints of the tasks in a hospital, a main focus has been devoted to the acceleration of the EM segmentation algorithm.
Progress
We developed 3 new features to improve the visualization of parameters in the EMSegment and to help the user select the optimum values:
- Intensity Normalization
- Intensity Distribution by 2D Gaussian
- Global Priors
Our development has been committed into the Slicer3 trunk and will be available in Slicer 3.5. As a side project, we developed a module for MRI Bias Field Correction: MRIBiasFieldCorrection We provided support to the NA-MIC group studying alcohol and stress interaction in primates, and we have tested the module on monkey datasets. Besides, a Command Line version of this module has been developed to enable automated Testing in Nightly builds.
New Features
Intensity Normalization
We compute the normalization parameter for the atlas based on the value selected by the user.
Intensity Distribution Visualization
We model the image intensities using a Gaussian distribution for every class.
Global Prior Visualization
We compute the Global Priors based on the selection in the intensity histogram.
Collaboration
- Bayesian Segmentation of MRI Images by Koen Van Leemput, Sylvain Jaume, Polina Golland, Steve Pieper and Ron Kikinis.
- NA-MIC NCBC Collaboration: Measuring Alcohol and Stress Interaction by Vidya Rajagopalan, Andriy Fedorov and Chris Wyatt.