Difference between revisions of "Engineering:GE"
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (19 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
Back to [[Engineering:Main|NA-MIC Engineering]] | Back to [[Engineering:Main|NA-MIC Engineering]] | ||
= Overview (PI: Jim Miller) = | = Overview (PI: Jim Miller) = | ||
| Line 20: | Line 21: | ||
| | | | | | ||
| − | == | + | == 3D Slicer Plugin Architecture == |
The 3D Slicer platform has been completely reworked through the joint | The 3D Slicer platform has been completely reworked through the joint | ||
efforts of the NA-MIC community and the [http://www.nbirn.net BIRN], | efforts of the NA-MIC community and the [http://www.nbirn.net BIRN], | ||
[http://nac.spl.harvard.edu NAC], [http://www.ncigt.org NCIGT], and | [http://nac.spl.harvard.edu NAC], [http://www.ncigt.org NCIGT], and | ||
| − | other cooperative grants that leverage the common infrastructure. GE Research has created a simple plugin architecture that allows researchers to extend | + | other cooperative grants that leverage the common infrastructure. GE Research has created a simple plugin architecture that allows researchers to extend Slicer's capabilities with plugins provided as executables, shared libraries, or python modules. See [https://www.slicer.org/wiki/Slicer3:Execution_Model_Documentation here] for more information. We have recently extended the execution model to include measurements either through [[2010 Winter Project Week Command Line Module Simple Return Types|Simple Return Parameters]] or tables of measurements in CSV (Comma Separated Value) files. Plugins can be built outside of the Slicer build tree and distributed as [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.2/SlicerApplication/ExtensionsManager Slicer Extensions]. Slicer plugins can shared with other applications, e.g. NiftyView and GIMIAS. See [[Media:AHM2010-Plug-ins.ppt|here]] and |
[http://www.nitrc.org/projects/slicer3examples/ here] for more information on interfacing with the 3D Slicer. | [http://www.nitrc.org/projects/slicer3examples/ here] for more information on interfacing with the 3D Slicer. | ||
|- | |- | ||
| − | | align="center" | [[Image: | + | | align="center" | [[Image:3DSlicerLogo-V-Color-201x204.png |150px|]] |
| | | | | | ||
| − | == | + | == Personal Slicer Cloud == |
| − | + | Leveraging the infrastructure of the 3D Slicer Plugin Architecture and the Remote IO mechanisms of Slicer, GE Research is developing a Personal Slicer Cloud. Personal Slicer Cloud allows a client machine to be paired with a server, allowing a user to schedule long running analyses on their server machine from their client machine for later review on their client machine. Here, the user will interact with the plugin on the client version of Slicer as they would normally but direct the module to run on the cloud instead of locally. The client version of Slicer will automatically package all the data and parameters necessary and submit to the user's Personal Slicer Cloud. A separate module on the client will allow the user to monitor the progress and download results back to the client. One use case would be to set up a module on your laptop, submit to your Personal Slicer Cloud, pack up your laptop for the drive home, and upon arriving at your destination open your laptop to download and review the results computed while you were in transit. | |
|- | |- | ||
| + | |||
|align="center" | [[Image:Annotated_Lightbox.png|300px|]] | |align="center" | [[Image:Annotated_Lightbox.png|300px|]] | ||
| Line 44: | Line 46: | ||
== Radiological Review Modes == | == Radiological Review Modes == | ||
| − | Clinical researchers expect image review workstations to provide a set of basic functionality. Handling DICOM as a source of image data is one example. Other examples include image multiplanar and oblique reformat, lightbox, comparisons, tracked cursors, ROI's, cine, and calipers for measurement. In conjunction with funding from [http://nac.spl.harvard.edu/ NAC], GE Research is providing some of these base capabilities to | + | Clinical researchers expect image review workstations to provide a set of basic functionality. Handling DICOM as a source of image data is one example. Other examples include image multiplanar and oblique reformat, lightbox, comparisons, tracked cursors, ROI's, cine, and calipers for measurement. In conjunction with funding from [http://nac.spl.harvard.edu/ NAC], GE Research is providing some of these base capabilities to the 3D Slicer. [https://www.slicer.org/wiki/Slicer3:RadiologicalReview More...] |
|- | |- | ||
| − | | align="center" | [[Image: | + | |align="center" | [[Image:Screen_Shot_2012-02-09_at_10.11.08_AM.png|250px|]] |
| | | | | | ||
| − | == | + | == Charting == |
| − | + | Quantitative imaging and longitudinal methods use charts to summarize and compare information between images. GE Research is building a charting infrastructure in the 3D Slicer which treats charts as a fundamental display component, akin to images and the 3D view. Charts can be packed in a layout with image viewers and 3D viewers, controlled directly from modules, constructed and modified interactively, serialized with the MRML scene, and emit signals to the application [https://www.slicer.org/wiki/Documentation/4.2/Developers/Charts More...] | |
| − | |- | + | |- |
| − | | align="center" | [[Image: | + | |align="center" | [[Image:AdditionalEdit.png|250px|]] |
| | | | | | ||
| − | == | + | == Interactive methods == |
| − | + | Interactive methods for segmentation and registration allow users of the 3D Slicer to label and align images through the users interactions for circumstances where automated methods are not practical, are not feasible, or are not necessary. GE Research has developed an architecture within the Editor module to support interactive segmentation, with a first embodiment being an implementation of the GrowCut method. This architecture provides semi-automated segmentation which allows the user to iterate and modify the segmentation. Current efforts include extending interactive methods to registration. [https://www.slicer.org/wiki/Modules:GrowCutSegmentation-Documentation-3.6 More...] | |
|- | |- | ||
| − | | align="center" | [[Image: | + | |align="center" | [[Image:PkModeling.png|75px|]] |
| | | | | | ||
| − | == | + | == Quantitative methods == |
| − | + | Pharmacokenetic Modeling (Pk Modeling) transforms Dynamic Contrast Enhance MRI (DCE MRI) imaging into quantitative measures of function. GE Research is working on a Slicer Extension and enabling Slicer architecture to support the Pk analysis of DCE MRI. | |
|- | |- | ||
| − | | align="center" | [[Image: | + | | align="center" | [[Image:HammerABrain.png|220px]] |
| | | | | | ||
| − | == | + | == Robust MRI Measurement Tools == |
| + | |||
| + | Funded through the NCBC collaboration grant mechanism, this project brings together researchers from [http://www.med.unc.edu/~dgshen/ UNC] and GE Research to develop deformable registration and MS lesion segmentation tools. The first generation of these tools are available as Slicer Extensions. [[NA-MIC_NCBC_Collaboration:Development_and_Dissemination_of_Robust_Brain_MRI_Measurement_Tools | More...]] | ||
| + | |||
| + | |- | ||
| − | |||
|} | |} | ||
Latest revision as of 17:11, 10 July 2017
Home < Engineering:GEBack to NA-MIC Engineering
Overview (PI: Jim Miller)
GE is adding technology to ITK to support the needs of NA-MIC, developing a module plugin and scheduling architecture for the 3D Slicer, developing standard radiological review modes, quantitative imaging techniques, charting, and interactive methods.
GE Research Projects

|
Insight Toolkit (ITK)The Insight Segmentation and Registration Toolkit (ITK) is an open-source software toolkit for performing registration and segmentation. ITK is used as the principal library for image analysis and general image IO in the 3D Slicer. GE Research has extended ITK to support NA-MIC activities by providing probability distributions, tensor pixels, level tracing, and displacement transforms. |

|
3D Slicer Plugin ArchitectureThe 3D Slicer platform has been completely reworked through the joint efforts of the NA-MIC community and the BIRN, NAC, NCIGT, and other cooperative grants that leverage the common infrastructure. GE Research has created a simple plugin architecture that allows researchers to extend Slicer's capabilities with plugins provided as executables, shared libraries, or python modules. See here for more information. We have recently extended the execution model to include measurements either through Simple Return Parameters or tables of measurements in CSV (Comma Separated Value) files. Plugins can be built outside of the Slicer build tree and distributed as Slicer Extensions. Slicer plugins can shared with other applications, e.g. NiftyView and GIMIAS. See here and here for more information on interfacing with the 3D Slicer. |

|
Personal Slicer CloudLeveraging the infrastructure of the 3D Slicer Plugin Architecture and the Remote IO mechanisms of Slicer, GE Research is developing a Personal Slicer Cloud. Personal Slicer Cloud allows a client machine to be paired with a server, allowing a user to schedule long running analyses on their server machine from their client machine for later review on their client machine. Here, the user will interact with the plugin on the client version of Slicer as they would normally but direct the module to run on the cloud instead of locally. The client version of Slicer will automatically package all the data and parameters necessary and submit to the user's Personal Slicer Cloud. A separate module on the client will allow the user to monitor the progress and download results back to the client. One use case would be to set up a module on your laptop, submit to your Personal Slicer Cloud, pack up your laptop for the drive home, and upon arriving at your destination open your laptop to download and review the results computed while you were in transit. |
|
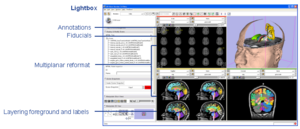
Radiological Review ModesClinical researchers expect image review workstations to provide a set of basic functionality. Handling DICOM as a source of image data is one example. Other examples include image multiplanar and oblique reformat, lightbox, comparisons, tracked cursors, ROI's, cine, and calipers for measurement. In conjunction with funding from NAC, GE Research is providing some of these base capabilities to the 3D Slicer. More... |
|
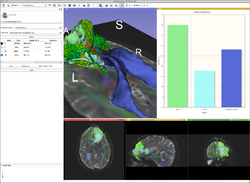
ChartingQuantitative imaging and longitudinal methods use charts to summarize and compare information between images. GE Research is building a charting infrastructure in the 3D Slicer which treats charts as a fundamental display component, akin to images and the 3D view. Charts can be packed in a layout with image viewers and 3D viewers, controlled directly from modules, constructed and modified interactively, serialized with the MRML scene, and emit signals to the application More... |

|
Interactive methodsInteractive methods for segmentation and registration allow users of the 3D Slicer to label and align images through the users interactions for circumstances where automated methods are not practical, are not feasible, or are not necessary. GE Research has developed an architecture within the Editor module to support interactive segmentation, with a first embodiment being an implementation of the GrowCut method. This architecture provides semi-automated segmentation which allows the user to iterate and modify the segmentation. Current efforts include extending interactive methods to registration. More... |

|
Quantitative methodsPharmacokenetic Modeling (Pk Modeling) transforms Dynamic Contrast Enhance MRI (DCE MRI) imaging into quantitative measures of function. GE Research is working on a Slicer Extension and enabling Slicer architecture to support the Pk analysis of DCE MRI. |
|
Robust MRI Measurement ToolsFunded through the NCBC collaboration grant mechanism, this project brings together researchers from UNC and GE Research to develop deformable registration and MS lesion segmentation tools. The first generation of these tools are available as Slicer Extensions. More... |
Prior GE Research Projects
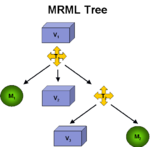
|
Registration ArchitectureA fundamental technology in Slicer is the scene graph or MRML scene used to describe the relationships between various sources of primary and derived data and annotation. Relating images from longitudinal or cross-sectional studies requires estimating the transformation between the image sets, a process referred to as registration. GE Research is providing an infrastructure for registration inside of Slicer3, leveraging existing registration algorithms from ITK, packaged as plugins that can be used inside or outside of Slicer3. The architecture includes IO mechanisms from transforms from files and plugins, transform representations in MRML, visualization through transforms, and managing the differences in coordinate frames between Slicer3 and ITK (see here and here). |
|
DICOM Support for Diffusion ImagingDiffusion weighted MR images in DICOM format are not currently recognized by Slicer as DW-MR images due to many proprietary tags and data formats used by different vendors. We created a plugin module for converting DICOM series of diffusion weighted images into NRRD volumes where correct image information (diffusion weighting directions and b-values) are parsed from the DICOM header and stored in the appropriate fileds in NRRD header. The converter supports some of the common formats from GE, Siemens, and Philips. Once the list of supported formats is reletively complete, the DICOM support for diffusion weighted images will be moved to itkVTKArchetypeIO. |

|
Skull Stripping and Tissue ClassificationSkull stripping and tissue classification as fundamental processes in a neuro imaging workflow. We are developing techniques which are well suited for the image exploration and discovery stages of interacting with a dataset. We are also collaborating with Jerry Prince's group from JHU and other interested parties within NA-MIC who are developing more detailed methods. The skull stripping algorithm we have developed is a parametric deformable model that evolves under the influence of the image intensity. The algorithm is designed to run quickly, making it amenable to these image exploration tasks. The algorithm has been tested on more than 300 OASIS datasets and compared against the BET results provided in the OASIS database, the average DICE score is 0.94. For the tissue classification, we implemented a fuzzy segmentation algorithm using ITK. The algorithm uses a quadratic polynomial to model the smooth gain field and simultaneously estimate/refine the polynomial parameters tissue classification. Both sets of algorithms are available as Slicer Extensions. |
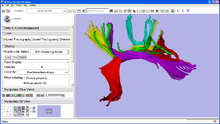
|
Quantitative analysis of fiber bundlesThrough a collaboration with the NAC, GE Research is developing a plugin for the 3D Slicer which clusters fiber tracts into anatomically meaningful fiber bundles and reports quantitative diffusion measures along the fiber bundles. This type of analysis has been applied to the study of Schizophrenia and in Neuro Surgical evaluations. The tract clustering and diffusion measurements are available as a Slicer Extension. See here for a detailed tutorial. |