Difference between revisions of "Events:RSNA 2009"
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | {| class="wikitable" | ||
| + | | colspan="3"|[[Image:RSNA2009.jpg]] | ||
| + | |- | ||
| + | | style="width:33%" |[[Image:NAMIC.jpg]] | ||
| + | | style="width:33%" |[[Image:Logo_nac.gif]] | ||
| + | |style="width:33%" |[[Image:NCIGTlogo.gif]] | ||
| + | |} | ||
| + | |||
[[File:RSNA2009 3DAnatomyCourse SPujol3.PNG | 400 px | right]] | [[File:RSNA2009 3DAnatomyCourse SPujol3.PNG | 400 px | right]] | ||
| Line 5: | Line 13: | ||
=Introduction= | =Introduction= | ||
The 3D Interactive Visualization of DICOM Images with Slicer course is offered by the National Alliance for Medical Image Computing (NA-MIC) in conjunction with the Neuroimage Analysis Center (NAC) at the [http://rsna2009.rsna.org/ 95th Scientific Assembly and Annual Meeting of the Radiological Society of North America (RSNA 2009)]. | The 3D Interactive Visualization of DICOM Images with Slicer course is offered by the National Alliance for Medical Image Computing (NA-MIC) in conjunction with the Neuroimage Analysis Center (NAC) at the [http://rsna2009.rsna.org/ 95th Scientific Assembly and Annual Meeting of the Radiological Society of North America (RSNA 2009)]. | ||
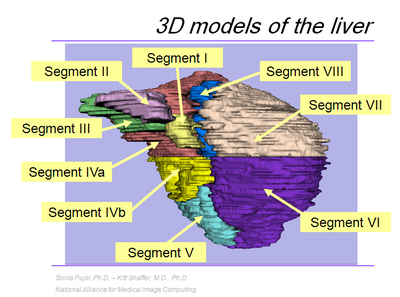
| − | As part of the outreach missions of these NIH funded National Centers, we have developed an offering of freely available, multi-platforms open source software to enable medical image analysis research. The [[Media:3DVisualization_SoniaPujol_RSNA2009_11-30-2009.pdf | course]] along with the tutorial [ | + | As part of the outreach missions of these NIH funded National Centers, we have developed an offering of freely available, multi-platforms open source software to enable medical image analysis research. The [[Media:3DVisualization_SoniaPujol_RSNA2009_11-30-2009.pdf | '''course''']] along with the tutorial [https://www.slicer.org/wiki/File:Slicer3VisualizationDataset.zip 3D Visualization] and [[Media:LiverData_RSNA2009.tar.gz | Liver]] datasets aim to introduce translational clinical scientists to the capabilities of the 3D Slicer software. There will be additional demos of Slicer capabilities at the [[Events:RSNA_CTSA_2009|CTSA event on Tuesday morning]]. |
=Target Audience= | =Target Audience= | ||
Latest revision as of 18:04, 10 July 2017
Home < Events:RSNA 2009
| ||

|

|

|
Contents
Introduction
The 3D Interactive Visualization of DICOM Images with Slicer course is offered by the National Alliance for Medical Image Computing (NA-MIC) in conjunction with the Neuroimage Analysis Center (NAC) at the 95th Scientific Assembly and Annual Meeting of the Radiological Society of North America (RSNA 2009). As part of the outreach missions of these NIH funded National Centers, we have developed an offering of freely available, multi-platforms open source software to enable medical image analysis research. The course along with the tutorial 3D Visualization and Liver datasets aim to introduce translational clinical scientists to the capabilities of the 3D Slicer software. There will be additional demos of Slicer capabilities at the CTSA event on Tuesday morning.
Target Audience
Radiology Residents and Fellows and others who are interested in medical image computing
Learning Objectives
The learning objectives of the workshop are (a) to enhance interpretation of DICOM images through the use of 3D visualization, (b) to gain experience with interactive assessment of complex anatomical structures (c) to present current directions of open-source computer graphics applications in Radiology. After completion of the workshop, participants will be able to visualize and analyze their own datasets within the 3DSlicer platform. Features that will be used include 3D Data Loading and Visualization, the Lightbox capabilities, Scene snapshots and Interactive Editing.
The course will include a tour of the DICOM/PACS resources available within the new Slicer3.5 software ,as well as an overview of training materials for advanced image analysis such as automatic segmentation of Brain MRI images, and Diffusion Tensor Imaging.
Faculty
- Sonia Pujol, Ph.D., Brigham and Women's Hospital, Harvard Medical School, Boston, MA
- Kitt Shaffer, M.D., Ph.D.,Boston University Medical Center, Harvard Medical School, Boston, MA
- Randy Gollub, M.D., Ph.D., Massachusetts General Hospital,Harvard Medical School, Boston, MA
- Kathryn Hayes, M.S.E, Brigham and Women's Hospital, Harvard Medical School, Boston, MA
- Ron Kikinis, M.D., Brigham and Women's Hospital, Harvard Medical School, Boston, MA
Logistics
- Date: Monday November 30, 2009
- Time: 2:30-4:00 pm
- Location: S401CD, McCormick Place, Chicago, Illinois
- This is "RSNA Informatics Course" IA21 and approved for AMA and PRA Category 1 Credit
- To register for the class, please sign up as a refresher course on the RSNA 2009 website.
Tentative Agenda
- 2:30 pm - 2:45 pm Slicer3 Overview and Applications (Sonia Pujol)
- 2:45 pm - 3:30 pm Hands-on Session 1: Data Loading and 3D Visualization (Sonia Pujol)
- 3:30 pm - 4:00 pm Hands-on Session 2: Using 3D Visualization to aid radiological readings: abdominal CT cases (Sonia Pujol/Kitt Shaffer)
Back to NA-MIC events