Home < Events:VT-NAMIC-NCBC-Collaboration-meeting-2009
Logistics
- Andriy Fedorov and Ginger Li: work on pre-meeting agenda Wed-Thu October 7-8, 2009
- All participants meeting: Friday, October 9, 2009, 9am-4pm (time is tentative, but room has been reserved, confirmed with Katie Mastrogiacomo)
- Wednesday and Thursday location: 1249 Boylston Street
- Friday meeting location: 1249 Boylston Street, 2nd floor conference room
Goals
- To be completed or partially completed prior to the meeting by Andriy Fedorov and Ginger Li (Oct 7-8):
- Provide necessary user- and developer-level Slicer training for Ginger
- Understand the currently used processing workflow
- Confirm reproducibility of the processing steps between VT and SPL
- Document the processing steps as precisely as necessary for reproducibility and cross-validation of the results (wiki page)
- To be completed during the meeting
- Review and confirm the correctness of the workflow based on the wiki documentation (Chris, Ron and Sandy)
- Review the available data and current segmentation results
- Discuss action timeline for the remaining part of the funded project period
- Potential for segmentation validation using cryo-histology (Chris can show some results of reconstructed 3D cryo-histo volumes)
- Potential for cortical thickness measures / integration with Caret or freesurfer
- Discuss publication plans
Agenda
- Review, discuss and finalize the processing workflow (to be discussed and documented by Andriy Fedorov and Ginger Li prior to the meeting Oct 7-8)
- atlas construction
- image pre-processing
- registration
- segmentation
- Review currents results
- Tommy and Louis images appear to have bad quality. Ron recommends not to use these subjects for atlas construction
- Identify Slicer components of the workflow that are ... working, broken, missing
Schedule
- Wednesday, Oct 7 plans:
- Walk through the segmentation workflow with EM Segmenter, verify scene saving, usability (Ginger)
- Document the current segmentation problems, prepare for discussion with Sandy
- 4pm: Meet with Sandy at Thorn to discuss current EM Segmenter results
- Install/verify installation of Slicer and related modules (BRAINSFit, N3MRILightCLI) (Andrey)
- Using and developing Slicer modules/extensions (Andrey)
- Using BRAINSFit (Andrey)
- AC-PC module usage for pose alignment (Ginger)
- Data transfer (images + segmentations)
- Aftenoon: Ron stops by to review progress (?)
- Friday, Oct 9, 9am (earlier?): Ron, Sandy, Ginger, Andrey meet at Boylston conference room to start discussion
- 10am: Chris' flight arrives at Logan
- 1pm: Chris gives a journal club talk at 1249 Boylston conference room
- 7pm: Chris' flight departs from Logan
Progress
- Oct 7
- Reviewed available data
- The issue with noise in Tommy and Louis resolved: we do not know how the NIFTI images we used before for Tommy and Louis were obtained -- the DICOM data has much better noise characteristics
We made the following observations about the data at the first time point:
- In some sobjects the FOV is axis-aligned, while in others it is tilted. These directions were not preserved in the NIFTI data we were previously using.
- We have two SPGR scans of for each subject, but the scans are not perfectly aligned. Ron: possibly due to coil heating. We cannot right away use these images to average and get better SNR, need to check with imaging people. Not sure if discrepancy is the rigid one.
- Noticeable bias field in the atlas is observed by Ron.
Other observations with illustrations:
| The issue with noise in Tommy and Louis resolved: we do not know how the NIFTI images we used before for Tommy and Louis were obtained -- the DICOM data has much better noise characteristics
|
|
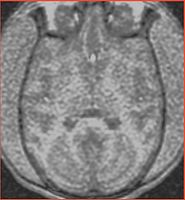
| Angio sequence (right name?) has apparently been used for some (7 out of 10) subjects
|
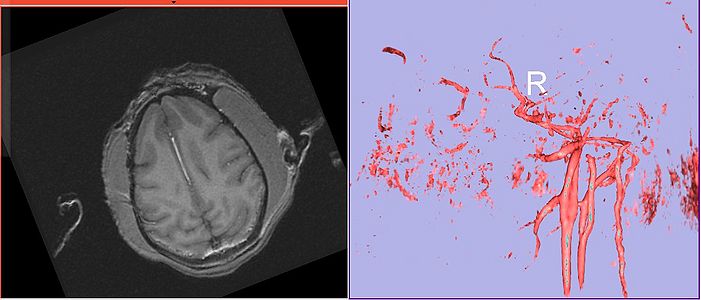
Calvin axial showing vessel and volume rendering of vasculature
|
|
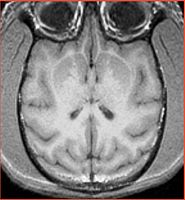
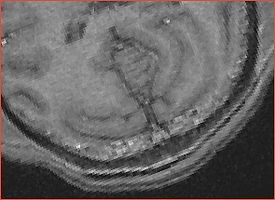
| Strange artifact is observed in the occipital lobe for all subjects. Ron, Sandy, Sota Oguro (SPL radiologist) cannot explain the origin. See Calvin example (note the interpolation is turned off)
|
|
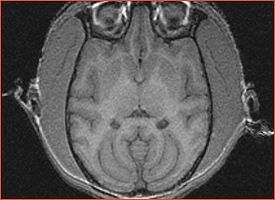
| Significant mis-alignment of some subjects with the template subject after FLIRT registration is confirmed
|
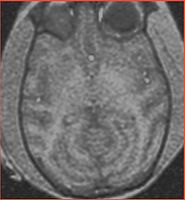
| Ringing artifacts for some (few) subjects
|
|
Attendance
Open to all interested parties:
- Chris Wyatt
- Ginger Li
- Ron Kikinis
- Sandy Wells
- Andriy Fedorov
References
- Measuring alcohol and stress interaction NCBC Collaboration page
- Atlas construction workflow, put together by Ginger prior to the meeting
- Slicer 2 vs. Slicer 3 EM Segmentation performance qualitative comparison, completed by Ginger prior to the meeting; also as a table
- Atlas construction workflow off BSL web site
- Evaluation of BRAINSFit performance on atlas-to-subject vervet registration, completed by Andrey prior to the meeting
- M. Styner, R. Knickmeyer, S. Joshi, C. Coe, S. J. Short, and J. Gilmore. Automatic brain segmentation in rhesus monkeys. Proc SPIE Medical Imaging Conference, Proc SPIE Vol 6512 Medical Imaging 2007, pp 65122L-1 - 65122L-8 pdf
- Revised atlas construction workflow proposal, completed by Andriy Fedorov, communicated to Ron, Sandy, Sylvain Jaume, discussed with Sandy, all prior to the meeting