Exploring multi-modal registration for improved longitudinal modeling of patient-specific 4D DTI data
From NAMIC Wiki
Home < Exploring multi-modal registration for improved longitudinal modeling of patient-specific 4D DTI data
Key Investigators
- Utah: Anuja Sharma, Bo Wang, Guido Gerig
- UCLA: Andrei Irimia, Micah Chambers, Jack Van Horn
Objective
- Assess possible white matter atrophy in TBI patients by utilizing multi-modal longitudinal information and quantify changes in white matter properties of tracts along time.
- Perform multi-modal image registration and utilize the lesion segmentation framework previously developed by Wang et al. for identifying tracts of interest.
- Collaborate closely with the UCLA team on this project.
Approach, Plan
- Co-register patient-specific structural and DTI data, together with lesion segmentations. Registration is performed using Slicer in a longitudinal fashion to establish correspondence between the acute and chronic scans from a patient across multiple modalities.
- Utilize information from the lesion segmentations to guide the tractography and identify tracts possibly exhibiting white matter atrophy.
- Assess white matter property changes longitudinally in tracts of interest.
Progress
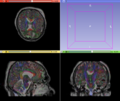
- Prototyping of the complete analysis pipeline using Slicer was successfully completed.
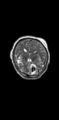
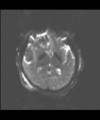
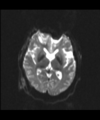
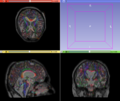
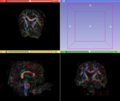
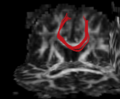
- Multi-modal registration for a TBI patient was done using Slicer. for the acute timepoint, the DTI scan was registered to the T2, via the B0 image. Similarly, for the followup chronic scan, T2 and DTI were registered to a common space. Following this, the registered DTI volumes from the two timepoints were co-registered using the derived FA images and resampling DTI using the estimated B-spline transform. Lesions estimated from structural MRI, FLAIR and GRE (Wang 2013) were also available in the same space as DTI owing to the multi-modal registration. The proximity to the lesion location prompted us to explore the mid-corpus callosum tract for possible white matter atrophy over time.
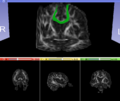
- Therefore, once all modalities were longitudinally registered, the mid-corpus callosum tract was obtained using label map seeding tractography and manually marked ROIs. Since the volumes were longitudinally co-registered, FA profiles of the mid-corpus callosum were then calculated and compared across acute and chronic timepoints.
- Closely collaborated with the UCLA team on this project. The initial Nifti to nrrd conversion of the dicom DWI data highlighted some loopholes in the Dicom converter utility and were worked upon by Micah. Discussed progress and future algorithm development of the above pipeline with Andrei.
- Other collaborations: Ron and Francois suggested possible options for alternate Slicer extensions and approaches for multi-modal and longitudinal DTI registration. Had discussions with Dominik on fine tuning the registration quality achieved during this week. This also initiated a request for a tutorial on DTI registration using Slicer. In the same light, Lauren suggested ways to clean up the original DWI's quality to further improve the accuracy of registration. Got inputs on new methods for white matter tract extraction by Demian.
References
- Bo Wang, Marcel Prastawa, Andrei Irimia, Micah C. Chambers, Neda Sadeghi, Paul M. Vespa, John D. Van Horn, Guido Gerig, Analyzing Imaging Biomarkers for Traumatic Brain Injury Using 4D Modeling of Longitudinal MRI, In IEEE ISBI 2013 .
- A. Sharma, P.T. Fletcher, J.H. Gilmore, M.L. Escolar, A. Gupta, M. Styner, G. Gerig. “Spatiotemporal Modeling of Discrete-Time Distribution-Valued Data Applied to DTI Tract Evolution in Infant Neurodevelopment,” In IEEE Proceedings of ISBI 2013.