Difference between revisions of "ITK Spherical Harmonics filter for shape analysis of cell nuclei"
From NAMIC Wiki
Rmachiraju (talk | contribs) |
|||
| (9 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
<gallery> | <gallery> | ||
Image:PW-MIT2010.png|[[2010_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2010.png|[[2010_Summer_Project_Week#Projects|Projects List]] | ||
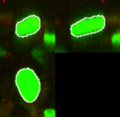
| + | Image:cell1.png|Cell Nucleus in the Tumor Microenvironment. | ||
| + | Image:cell2.png|SPHARM-PDM of nucleus. | ||
</gallery> | </gallery> | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * OSU: | + | * OSU: Shantanu Singh, Raghu Machiraju |
| − | * Harvard Medical School: Sean Megason | + | * Harvard Medical School: Arnaud Gelas, Kishore Mosaliganti, Sean Megason |
==Project== | ==Project== | ||
| Line 14: | Line 16: | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | ||
| − | + | * In our recent work[1] on studying the variation of nuclear structure in tumor microenvironments [2], we have used SPHARM-PDM [3] to model 3D nuclear shape. We are interested in using itkQuadEdgeMesh as the mesh representation to this end and will be evaluating the use of this class for our system. | |
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| Line 25: | Line 23: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | * Use implementation of SPHARM-PDM [3] as a starting point | |
| − | + | * Evaluate efficacy of itkQuadEdgeMesh to compute other descriptors [4] of nuclear surface | |
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| Line 35: | Line 30: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | * Current system for statistical shape analysis of nuclear morphology uses [3] to generate the representation | |
| − | + | ||
| − | + | * With Arnaud Gelas, initiated work on using itkQuadEdgeMesh for generating and representing PDM for nuclei | |
| − | + | * With Manasi Datar, initiated experiments on using particle based correspondence for generating PDMs | |
| + | * With Marco Ruiz, used GWE to set up the PDM generation on a large dataset of nuclei | ||
</div> | </div> | ||
| + | |||
</div> | </div> | ||
<div style="width: 97%; float: left;"> | <div style="width: 97%; float: left;"> | ||
| + | |||
| + | 1. Shantanu Singh, Sundaresan Raman, Enrico Caserta, Gustavo Leone, Michael Ostrowski, Jens Rittscher, Raghu Machiraju: Analysis of Spatial Variation of Nuclear Morphology in Tissue Microenvironments, 7th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2010. | ||
| + | |||
| + | 2. Trimboli et. al., Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature 461 (7267) p. 1084-91, 2009 | ||
| + | |||
| + | 3. Martin Styner, Ipek Oguz, Shun Xu, James J Levitt, Martha E Shenton, Guido Gerig Framework for the Statistical Shape Analysis of Brain Structures using SPHARM-PDM, Insight Journal p. 1-20 (2006) | ||
Latest revision as of 13:54, 25 June 2010
Home < ITK Spherical Harmonics filter for shape analysis of cell nucleiKey Investigators
- OSU: Shantanu Singh, Raghu Machiraju
- Harvard Medical School: Arnaud Gelas, Kishore Mosaliganti, Sean Megason
Project
Objective
- In our recent work[1] on studying the variation of nuclear structure in tumor microenvironments [2], we have used SPHARM-PDM [3] to model 3D nuclear shape. We are interested in using itkQuadEdgeMesh as the mesh representation to this end and will be evaluating the use of this class for our system.
Approach, Plan
- Use implementation of SPHARM-PDM [3] as a starting point
- Evaluate efficacy of itkQuadEdgeMesh to compute other descriptors [4] of nuclear surface
Progress
- Current system for statistical shape analysis of nuclear morphology uses [3] to generate the representation
- With Arnaud Gelas, initiated work on using itkQuadEdgeMesh for generating and representing PDM for nuclei
- With Manasi Datar, initiated experiments on using particle based correspondence for generating PDMs
- With Marco Ruiz, used GWE to set up the PDM generation on a large dataset of nuclei
1. Shantanu Singh, Sundaresan Raman, Enrico Caserta, Gustavo Leone, Michael Ostrowski, Jens Rittscher, Raghu Machiraju: Analysis of Spatial Variation of Nuclear Morphology in Tissue Microenvironments, 7th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, 2010.
2. Trimboli et. al., Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature 461 (7267) p. 1084-91, 2009
3. Martin Styner, Ipek Oguz, Shun Xu, James J Levitt, Martha E Shenton, Guido Gerig Framework for the Statistical Shape Analysis of Brain Structures using SPHARM-PDM, Insight Journal p. 1-20 (2006)