Difference between revisions of "Imaging in Neuroscience"
From NAMIC Wiki
(→Agenda) |
(→Agenda) |
||
| Line 22: | Line 22: | ||
<font color=red>Note: | <font color=red>Note: | ||
| − | * Slide page 37: "Load scene" menu is not available in the current version of 3D Slicer (version 4.2.2). Please choose "Add data" instead. | + | * Slide page 37 (For those who downloaded the slide above before Sunday): "Load scene" menu is not available in the current version of 3D Slicer (version 4.2.2). Please choose "Add data" instead. |
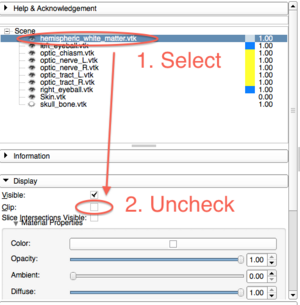
* Slice page 48: If the white matter surface does not appear, please select "hemisphelic_white_matter.vtk" and uncheck "Clip" (see the screenshot bellow). | * Slice page 48: If the white matter surface does not appear, please select "hemisphelic_white_matter.vtk" and uncheck "Clip" (see the screenshot bellow). | ||
[[Image:ImagingInNeuroscience_April8_Clipping.png|300px]] | [[Image:ImagingInNeuroscience_April8_Clipping.png|300px]] | ||
Revision as of 18:05, 7 April 2013
Home < Imaging in NeuroscienceThe 3D Slicer software platform will be presented as part of the Harvard Catalyst course "Advanced Imaging: Imaging in Neuroscience" course organized by Dr. Randy Gollub
Course Faculty
- Junichi Tokuda, Brigham and Women's Hospital, Harvard Medical School (Presenter)
- Nicole Aucoin, Brigham and Women's Hospital, Harvard Medical School
- Sonia Pujol, Brigham and Women's Hospital, Harvard Medical School
- Randy L. Gollub, Massachusetts General Hospital, Harvard Medical School
- P. Ellen Grant, Boston Children's Hospital, Harvard Medical School
- Daniel Haehn, Boston Children's Hospital, Harvard Medical School
- Nicolas Rannou, Boston Children's Hospital, Harvard Medical School
- Nathaniel A Reynolds, Massachusetts General Hospital, Harvard Medical School
Course Logistics
Date: April 8, 2013, 11:30-12:00 Location: Sheraton Commander Hotel, Boston, MA
Agenda
- 11:30-12:00: The 3D Slicer open-source platform for image analysis and 3D visualization of medical image data (lecture)
- 12:00-1:00: Slicer4 minute tutorial (hands-on session) and Diffusion tractography (demo)
3D Slicer course - Imaging in Neuroscience
Note:
- Slide page 37 (For those who downloaded the slide above before Sunday): "Load scene" menu is not available in the current version of 3D Slicer (version 4.2.2). Please choose "Add data" instead.
- Slice page 48: If the white matter surface does not appear, please select "hemisphelic_white_matter.vtk" and uncheck "Clip" (see the screenshot bellow).
Course Materials
In preparation for the hands-on session, please download the 3D Slicer release version 4.2.2-1 and the Slicer4 minute datasets.
3D Slicer 4.x (blue download button)
Slicer4minute dataset (hands-on)
Additional materials for learning more about 3D Slicer are available on the Slicer 101 page.