Difference between revisions of "Loading and segmentation of histopathology imaging for radiological-pathological correlation"
| (6 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
Image:PW-MIT2012.png|[[2012_Summer_Project_Week#Projects|Projects List]] | Image:PW-MIT2012.png|[[2012_Summer_Project_Week#Projects|Projects List]] | ||
Image:histopath.png | Image:histopath.png | ||
| + | Image:Labelmap_histo.png | ||
</gallery> | </gallery> | ||
| Line 8: | Line 9: | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * SPL: Andrey Fedorov | + | * SPL: Tobias Penzkofer, Andrey Fedorov |
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 20: | Line 21: | ||
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | Find, analyze and adapt the routines responsible for 3 channel image handling to enable processing of scanned histology slides with possible extensions towards registration of histo/path, digital pathology imaging and | + | Find, analyze and adapt the routines responsible for 3 channel image handling to enable processing of scanned histology slides with possible extensions towards registration of histo/path, digital pathology imaging and analysis. |
| + | |||
| + | Specifically we would like to be able: | ||
| + | * loading / saving of Slicer scenes that contain RGB volumes | ||
| + | * masking and segmentation of RGB volumes in Editor | ||
</div> | </div> | ||
<div style="width: 40%; float: left;"> | <div style="width: 40%; float: left;"> | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | Possible solutions have been identified (Downsampling, NRRD file format save) and applied. | + | Possible solutions have been identified (DCM Convert, Downsampling, NRRD file format save) and applied. Reasons for Slicer not supporting Vector Data in the Editor have been identified (Threshold algorithm). Slicer native import algorithm can provide a solution for many problems (if fixed). |
| + | |||
| + | Bug report submitted: http://www.na-mic.org/Bug/view.php?id=2225 (current Slicer does not correctly load and display PNG image date) (FIXED by Julien) | ||
| + | |||
| + | Crash reading any DICOM scalar volume or PNG stack: http://www.na-mic.org/Bug/view.php?id=2241 (communicated to Demian) | ||
| + | |||
| + | Solution with secondary scalar volume works well (see image). | ||
| + | |||
</div> | </div> | ||
</div> | </div> | ||
| Line 36: | Line 48: | ||
##Built-in YES | ##Built-in YES | ||
##Extension -- commandline NO | ##Extension -- commandline NO | ||
| − | ##Extension -- loadable | + | ##Extension -- loadable NO |
#Other (Please specify) | #Other (Please specify) | ||
Latest revision as of 13:46, 22 June 2012
Home < Loading and segmentation of histopathology imaging for radiological-pathological correlationInstructions for Use of this Template
Key Investigators
- SPL: Tobias Penzkofer, Andrey Fedorov
Objective
To enable Slicer 4 to load, view and process histopathology data.
Approach, Plan
Find, analyze and adapt the routines responsible for 3 channel image handling to enable processing of scanned histology slides with possible extensions towards registration of histo/path, digital pathology imaging and analysis.
Specifically we would like to be able:
- loading / saving of Slicer scenes that contain RGB volumes
- masking and segmentation of RGB volumes in Editor
Progress
Possible solutions have been identified (DCM Convert, Downsampling, NRRD file format save) and applied. Reasons for Slicer not supporting Vector Data in the Editor have been identified (Threshold algorithm). Slicer native import algorithm can provide a solution for many problems (if fixed).
Bug report submitted: http://www.na-mic.org/Bug/view.php?id=2225 (current Slicer does not correctly load and display PNG image date) (FIXED by Julien)
Crash reading any DICOM scalar volume or PNG stack: http://www.na-mic.org/Bug/view.php?id=2241 (communicated to Demian)
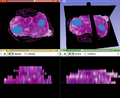
Solution with secondary scalar volume works well (see image).
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as a (please select the appropriate options by noting YES against them below)
- Slicer Module
- Built-in YES
- Extension -- commandline NO
- Extension -- loadable NO
- Other (Please specify)
References
- Trivedi, H., B. Turkbey, et al. (2012). "Use of patient-‐specific MRI-‐based prostate mold for validation of multiparametric MRI in localization of prostate cancer." Urology 79(1): 233-‐239.
- "Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information." Med Phys 38(4): 2005-‐2018.
- "Semi-‐automatic deformable registration of prostate MR images to pathological slices." J Magn Reson Imaging 32(5): 1149-‐1157.