Difference between revisions of "LupusLesionTutorial"
From NAMIC Wiki
Hjbockholt (talk | contribs) |
Hjbockholt (talk | contribs) m (→Tutorials) |
||
| Line 13: | Line 13: | ||
==Tutorials== | ==Tutorials== | ||
| − | • ''' | + | • '''Lupus Lesion tutorial''' : end-to-end Slicer3 module to perform automatic white matter lesion analysis[[Media:LupusLesion-Slicer3-Tutorial.ppt| [ppt]]] |
==Software== | ==Software== | ||
Revision as of 05:55, 7 January 2009
Home < LupusLesionTutorialContents
Lupus Lesion Slicer3 Tutorial
Overview
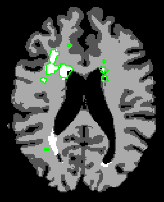
The following tutorial will allow you to perform an individual analysis of white matter lesions:
- Automatic analysis: in the lupus tutorial, you will learn how to load input volumes, run the end-to-end module lesion analysis to tissue classify and display output lesion maps.
Roadmap project : http://wiki.na-mic.org/Wiki/index.php/DBP2:MIND:Roadmap
Tutorials
• Lupus Lesion tutorial : end-to-end Slicer3 module to perform automatic white matter lesion analysis [ppt]
Software
• Slicer3 http://www.slicer.org/pages/Downloads
Tutorial materials
cvs -d :pserver:anonymous@www.nitrc.org:/cvsroot/lupuslesion login
cvs -d :pserver:anonymous@www.nitrc.org:/cvsroot/lupuslesion checkout lupuslesion
cvs -d :pserver:anonymous@www.nitrc.org:/cvsroot/lupuslesion checkout data
People
H Jeremy Bockholt
Mark Scully