Difference between revisions of "MedianTexture"
| (2 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<gallery> | <gallery> | ||
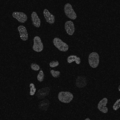
| − | Image: | + | Image:2TS0005_stack0001frame0005.png|Original PCNA-GFP fluorescence confocal microscope image for cell cycle marker studies. |
| − | Image: | + | Image:FGPAC_mask_101iter_stack0001frame0005.png|Multiphase GPAC segmentation mask. |
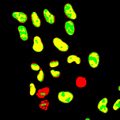
| − | Image: | + | Image:Masked-bkg mbp+2.png|Distribution of MBPs shown with each pattern mapped to a unique color. |
</gallery> | </gallery> | ||
| − | + | <gallery> | |
| − | + | Image:MBP_2TS0005_5U.png|MBP Uniform pattern 5. | |
| − | + | Image:MBP_2TS0005_6U.png|MBP Uniform pattern 6. | |
| − | + | Image:MBP_2TS0005_7U.png|MBP Uniform pattern 7. | |
| − | + | </gallery> | |
| − | + | <gallery> | |
| − | + | Image:MBP_2TS0005_8U.png|MBP Uniform pattern 8. | |
| + | Image:MBP_2TS0005_13U.png|MBP Uniform pattern 13. | ||
| + | Image:MBP_2TS0005_14U.png|MBP Uniform pattern 14. | ||
| + | </gallery> | ||
| + | <gallery> | ||
| + | Image:MBP_2TS0005_15U.png|MBP Uniform pattern 15. | ||
| + | Image:MBP_2TS0005_16U.png|MBP Uniform pattern 16. | ||
| + | Image:MBP_2TS0005_17U.png|MBP Uniform pattern 17. | ||
| + | </gallery> | ||
| + | <gallery> | ||
| + | Image:Mask_2TS0005_-1NU.png|MBP Non-Uniform pattern class. | ||
| + | </gallery> | ||
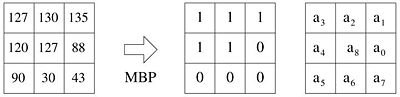
| + | [[File:MBP-definition.jpg|400px|thumb|left|Definition of MBP]] | ||
==Key Investigators== | ==Key Investigators== | ||
| − | * | + | * ENSI-Bourges, France: Lucas Menand, Sarah Portugais, Adel Hafiane |
| − | * | + | * Air Force Research Lab: Guna Seetharaman |
| + | * University of Missouri: Filiz Bunyak, K. Palaniappan | ||
| + | * Harvard: Sean Megason | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| − | <div style="width: | + | <div style="width: 40%; float: left; padding-right: 3%;"> |
<h3>Objective</h3> | <h3>Objective</h3> | ||
| − | + | Median binary patterns (MBP) are robust feature descriptors for characterizing natural and biological textures. MBPs can be used for cell segmentation, characterizing nuclei and membrane textures and for cell classification. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| − | <div style="width: | + | <div style="width: 40%; float: left; padding-right: 3%;"> |
<h3>Approach, Plan</h3> | <h3>Approach, Plan</h3> | ||
| − | + | MBPs is a robust alternative to the local binary pattern (LBP) texture descriptors that uses the local median instead of the central pixel intensity as the reference value to create a binary pattern. MBPs (and LBPs) have the attractive properties of noise-resistance, rotation invariance and shift-invariance and provide a powerful feature set for cell segmentation and classification. Using a 3x3 median window there are nine special median uniform patterns and a set of non-uniform patterns that we assign to a separate category. | |
| − | |||
| − | |||
| − | |||
</div> | </div> | ||
| Line 43: | Line 49: | ||
<h3>Progress</h3> | <h3>Progress</h3> | ||
| − | + | We have completed C++ and Matlab implementation of 2D median binary patterns and have a preliminary ITK version that needs to be tested and evaluated for correctness. | |
| + | </div> | ||
| + | |||
| + | <h3>NA-MIC Week Progress</h3> | ||
| + | * Completed ITK implementation of MBP. | ||
| + | * Histograms of MBPs can be plotted using VTK widgets. | ||
| + | * Remaining work is to build test cases for ITK and write a ITK Journal article. | ||
| − | |||
</div> | </div> | ||
| Line 53: | Line 64: | ||
==Delivery Mechanism== | ==Delivery Mechanism== | ||
| − | This work will be delivered to the NA-MIC Kit as | + | This work will be delivered to the NA-MIC Kit as an: |
| − | |||
#ITK Module | #ITK Module | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==References== | ==References== | ||
| − | * | + | *A. Hafiane, G. Seetharaman, K. Palaniappan, B. Zavidovique, “Rotationally invariant hashing of median patterns for texture classification”, Lecture Notes in Computer Science (ICIAR), Vol. 5112, 2008, pp. 619-629. <http://www.ncbi.nlm.nih.gov/pubmed/19116672> |
| − | + | ||
| − | * | + | *A. Hafiane, Seetharaman, G., Zavidovique, B.: Median binary pattern for textures classification. Lecture Notes in Computer Science CIAR, 2007, pp. 387–398. |
| − | * | + | |
| + | *T. Ojala, Pietikainen, M., Maenpaa, T.: Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Transactions on Pattern Analysis and Machine Intelligence 24(7), 2002, pp. 971–987. | ||
</div> | </div> | ||
Latest revision as of 23:32, 24 June 2010
Home < MedianTextureKey Investigators
- ENSI-Bourges, France: Lucas Menand, Sarah Portugais, Adel Hafiane
- Air Force Research Lab: Guna Seetharaman
- University of Missouri: Filiz Bunyak, K. Palaniappan
- Harvard: Sean Megason
Objective
Median binary patterns (MBP) are robust feature descriptors for characterizing natural and biological textures. MBPs can be used for cell segmentation, characterizing nuclei and membrane textures and for cell classification.
Approach, Plan
MBPs is a robust alternative to the local binary pattern (LBP) texture descriptors that uses the local median instead of the central pixel intensity as the reference value to create a binary pattern. MBPs (and LBPs) have the attractive properties of noise-resistance, rotation invariance and shift-invariance and provide a powerful feature set for cell segmentation and classification. Using a 3x3 median window there are nine special median uniform patterns and a set of non-uniform patterns that we assign to a separate category.
Progress
We have completed C++ and Matlab implementation of 2D median binary patterns and have a preliminary ITK version that needs to be tested and evaluated for correctness.
NA-MIC Week Progress
- Completed ITK implementation of MBP.
- Histograms of MBPs can be plotted using VTK widgets.
- Remaining work is to build test cases for ITK and write a ITK Journal article.
Delivery Mechanism
This work will be delivered to the NA-MIC Kit as an:
- ITK Module
References
- A. Hafiane, G. Seetharaman, K. Palaniappan, B. Zavidovique, “Rotationally invariant hashing of median patterns for texture classification”, Lecture Notes in Computer Science (ICIAR), Vol. 5112, 2008, pp. 619-629. <http://www.ncbi.nlm.nih.gov/pubmed/19116672>
- A. Hafiane, Seetharaman, G., Zavidovique, B.: Median binary pattern for textures classification. Lecture Notes in Computer Science CIAR, 2007, pp. 387–398.
- T. Ojala, Pietikainen, M., Maenpaa, T.: Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Transactions on Pattern Analysis and Machine Intelligence 24(7), 2002, pp. 971–987.