Difference between revisions of "MeningiomaMRIRegistrationStudy"
| (11 intermediate revisions by the same user not shown) | |||
| Line 80: | Line 80: | ||
* qualitative assessment: visually pleasing results | * qualitative assessment: visually pleasing results | ||
| − | * quantitative assessment: | + | * quantitative assessment: ''relative'' Target Registration Error (TRE) statistics collected per-pixel over the segmented tumor volume. In the absence of ground truth for registration result, and considering the wide acceptance and thorough testing of FSL FLIRT registration tool, we accept its result to be optimal under existing operating conditions. We then evaluate each of the registration tools as follows: |
| + | *# Let T_{flirt} and T_{reg} be the transforms aligning the images being registered recovered with FSL FLIRT and the compared registration tool, respectively | ||
| + | *# Let T(p) be the coordinates of a point after applying a transform | ||
| + | *# We then define the relative registration error ''e'' as follows: ''e = ||T_{flirt}(p)-T_{reg}(p)||'' | ||
| + | *# Given the binary mask defining the tumor, we evaluate the value of registration error surrogate at each pixel corresponding to the tumor volume, and report the values of min, max, and average relative error values for this region. | ||
* minimum execution time to meet the application requirements: interactive quantification of tumor growth | * minimum execution time to meet the application requirements: interactive quantification of tumor growth | ||
* minimum tuning of registration parameters (is this possible? to answer this sub-question we compare the results with FLIRT registration using its default parameters) | * minimum tuning of registration parameters (is this possible? to answer this sub-question we compare the results with FLIRT registration using its default parameters) | ||
| Line 117: | Line 121: | ||
${FIXED_IMAGE} ${MOVING_IMAGE} | ${FIXED_IMAGE} ${MOVING_IMAGE} | ||
</pre> | </pre> | ||
| + | |||
| + | NOTE: the default value for the number of samples was increased by 10, and the number of histogram bins was increased to 50 from 30. | ||
* execution time: ~4min | * execution time: ~4min | ||
| Line 165: | Line 171: | ||
=Results= | =Results= | ||
| − | ==BRAINSFit== | + | ==Qualitative assessment== |
| + | |||
| + | ===BRAINSFit=== | ||
| + | |||
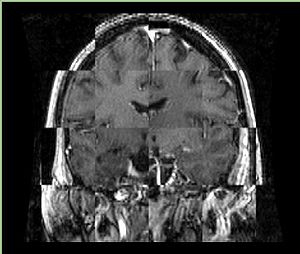
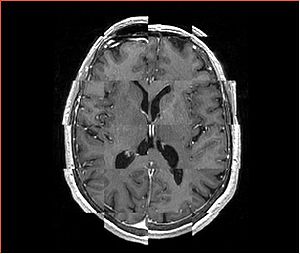
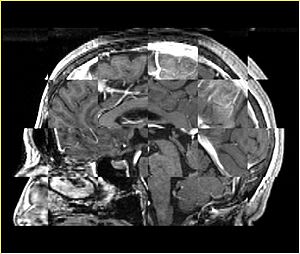
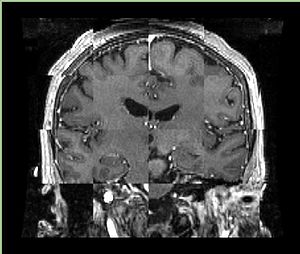
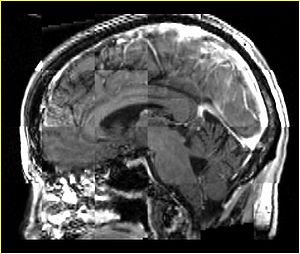
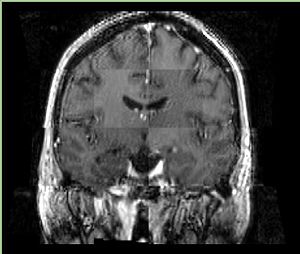
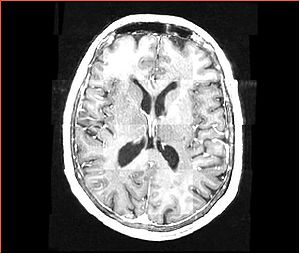
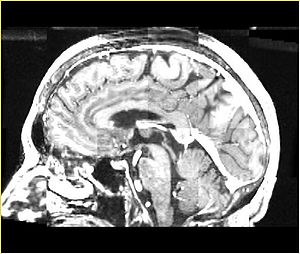
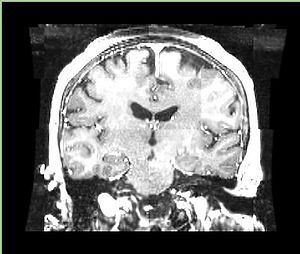
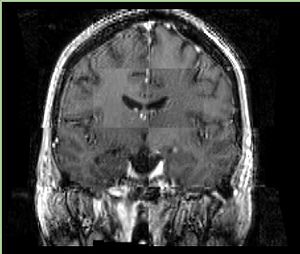
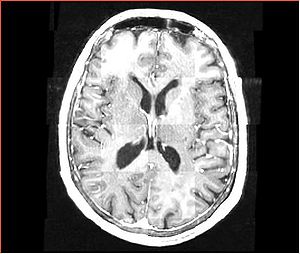
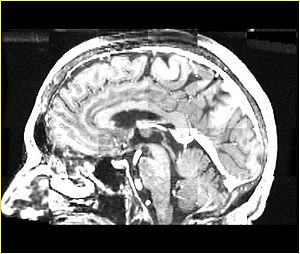
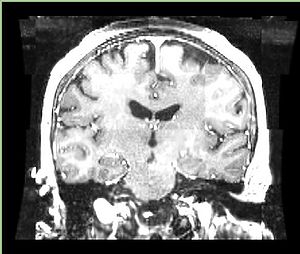
| + | A distinctive feature of BRAINSFit tool is the ability to automatically detect head region of interest, which is used for collecting intensity samples during metric calculation. Below is the overlay of such ROI in light blue overlayed with the image for Case10. This functionality appears to work consistently for all the cases used in the comparison. | ||
| + | |||
| + | <gallery perrow="3" heights="300px" widths="300px"> | ||
| + | Image:Case10-BF-ROI-a.jpg | ||
| + | Image:Case10-BF-ROI-s.jpg | ||
| + | Image:Case10-BF-ROI-c.jpg | ||
| + | </gallery> | ||
** Case01 | ** Case01 | ||
| Line 195: | Line 211: | ||
</gallery> | </gallery> | ||
| − | + | ===FSL FLIRT=== | |
| − | ==FSL FLIRT== | ||
** Case01 | ** Case01 | ||
| Line 225: | Line 240: | ||
Image:Case10-FLIRT-coronal.jpg | Image:Case10-FLIRT-coronal.jpg | ||
</gallery> | </gallery> | ||
| + | |||
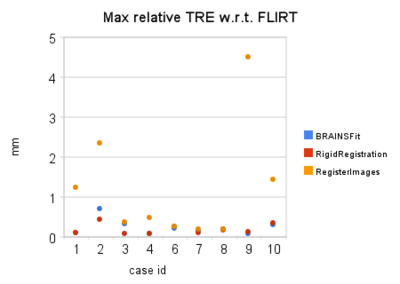
| + | ==Quantitative comparison== | ||
| + | <gallery perrow="3" heights="300px" widths="400px"> | ||
| + | Image:min_reg_tre_wrt_flirt.jpg | ||
| + | Image:ave_reg_tre_wrt_flirt.jpg | ||
| + | Image:max_reg_tre_wrt_flirt.jpg | ||
| + | </gallery> | ||
| + | |||
| + | Execution times recap (UPDATE: these times are for the code compiled in debug mode! In the release mode, execution time of BRAINSFit is under 1 minute): | ||
| + | |||
| + | * FLIRT: ~ 4 min | ||
| + | * BRAINSFit: > 10 min (with most of the time, ~80%, spent in ROI detection and transform initialization) | ||
| + | * RigidRegistration: ~ 4 min | ||
| + | * RegisterImages: ~ 2 min | ||
=Conclusions= | =Conclusions= | ||
| Line 242: | Line 271: | ||
* all of the tools implemented in Slicer require to specify the max number of iterations, which essentially requires the user to monitor convergence (metric value while iterating can be found in Slicer error log), and increase the number of iterations if the satisfactory result is not achieved. Not practical for interactive application | * all of the tools implemented in Slicer require to specify the max number of iterations, which essentially requires the user to monitor convergence (metric value while iterating can be found in Slicer error log), and increase the number of iterations if the satisfactory result is not achieved. Not practical for interactive application | ||
| − | * not possible to specify the similarity metric: apparently, FLIRT is able to get good result using cross correlation | + | * not possible to specify the similarity metric: apparently, FLIRT is able to get good result using cross correlation, which is less demanding computationally |
| + | |||
| + | * RegisterImages and RigidRegistration do not allow to specify 9-dof transforms, which are reportedly used to adjust for voxel size inconsistency in longitudinal studies [1]. | ||
| + | |||
| + | =Acknowledgements= | ||
| + | |||
| + | We acknowledge and appreciate the efforts of Hans Johnson, Jim Miller and Stephen Aylward in developing open source registration tools analyzed here. FSL FLIRT tool was used here solely for academic research purposes. We thank Luke Bloy for providing the code to perform conversion of FLIRT transform matrix into the ITK-readable format. | ||
| + | |||
| + | =References= | ||
| + | |||
| + | # J. L. Whitwell, J. M. Schotta, E. B. Lewis, D. G. Macmanus, and N. C. Fox, "Using nine degrees-of-freedom registration to correct for changes in voxel size in serial mri studies," Magnetic Resonance Imaging, vol. 22, no. 7, pp. 993-999, September 2004. [http://dx.doi.org/10.1016/j.mri.2004.01.057] | ||
Latest revision as of 20:02, 3 September 2009
Home < MeningiomaMRIRegistrationStudyContents
Objective
Accurate registration of same patient/same modality MRI data for longitudinal analysis of tumor progression.
Specific Aims
- Compare the accuracy of registration using existing Slicer and non-Slicer tools
- Identify parameter settings that produce satisfactory results
- Outline the limitations of the available registration tools in the context of the specific clinical research application
Data
- Input images: isotropic post-contrast T1 MRI acquired at different locations of BWH during 2006-2008, used under medical records study IRB. Time period between acquisition of scans for each patient is about 1 year.
- images were acquired with the same sequence, but possible on different scanners, all scans are axial
- Ground truth transformation: not available
- Expert landmarks for registration evaluation: not available
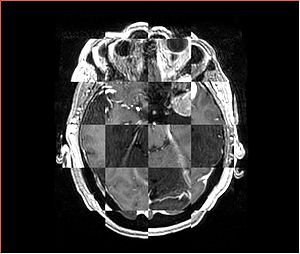
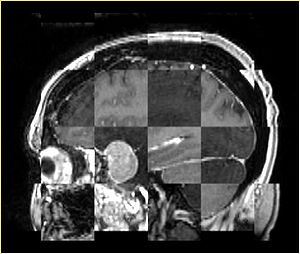
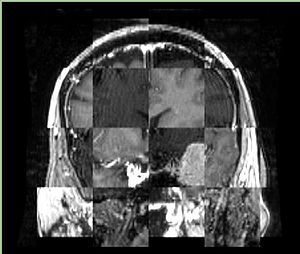
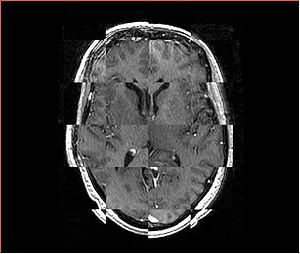
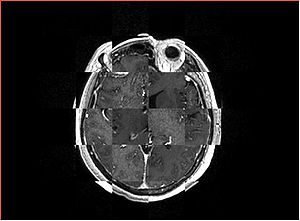
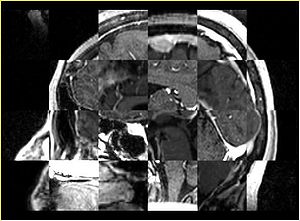
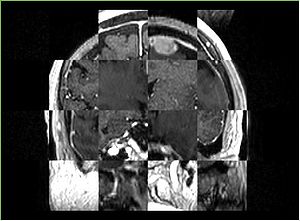
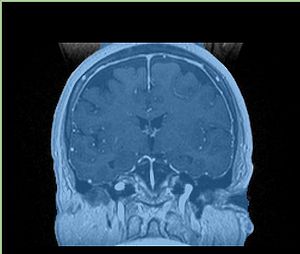
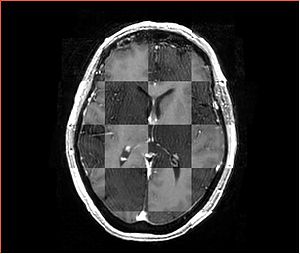
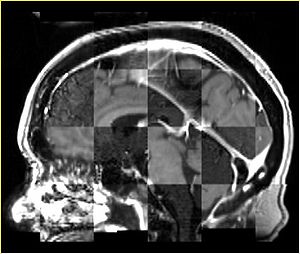
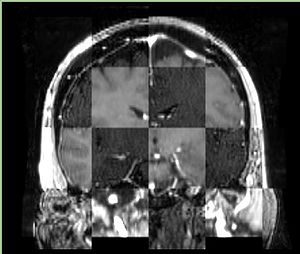
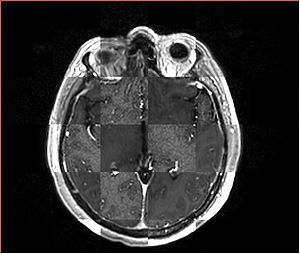
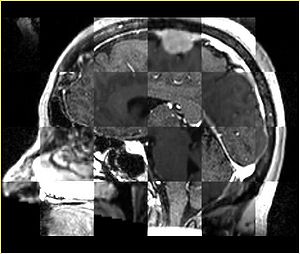
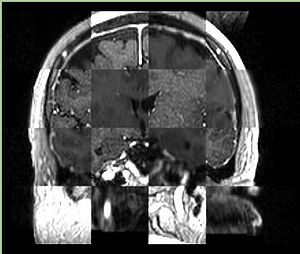
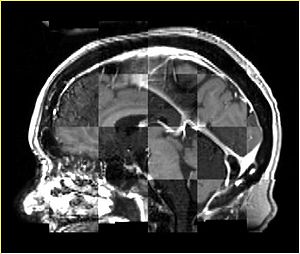
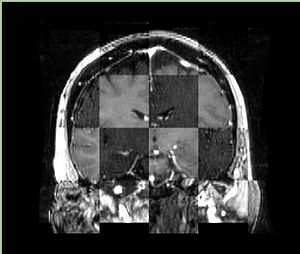
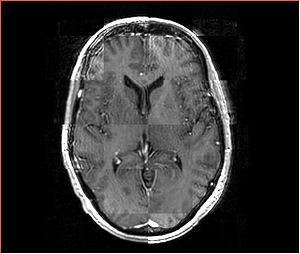
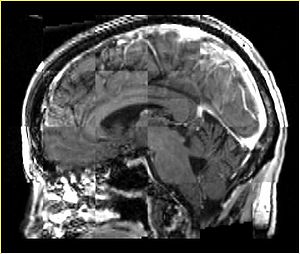
- Checkerboard appearance of unregistered images for the representative data of interest
- Case01
- Case03
- Case04
- Case10
Environment
- all registration methods were executed on george.bwh.harvard.edu, which has 16 cores like this one (output of "cat /proc/cpuinfo"):
processor : 0 vendor_id : AuthenticAMD cpu family : 15 model : 33 model name : Dual Core AMD Opteron(tm) Processor 880 stepping : 2 cpu MHz : 1000.000 cache size : 1024 KB physical id : 8 siblings : 2 core id : 0 cpu cores : 2 fpu : yes fpu_exception : yes cpuid level : 1 wp : yes flags : fpu vme de pse tsc msr pae mce cx8 apic sep mtrr pge mca cmov pat pse36 clflush mmx fxsr sse sse2 ht syscall nx mmxext fxsr_opt lm 3dnowext 3dnow rep_good pni lahf_lm cmp_legacy bogomips : 2010.02 TLB size : 1024 4K pages clflush size : 64 cache_alignment : 64 address sizes : 40 bits physical, 48 bits virtual power management: ts fid vid ttp
- Where number of threads was possible to specify (RegisterImages and BRAINSFit), it was set to 4. RigidRegistration uses maximum number of threads available, while FLIRT is single-threaded (need to be checked)
Measures of success
- qualitative assessment: visually pleasing results
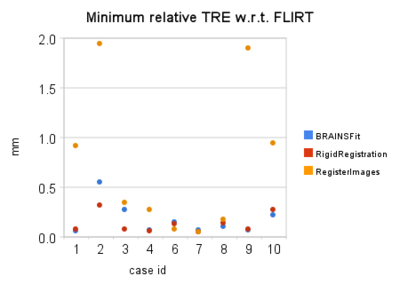
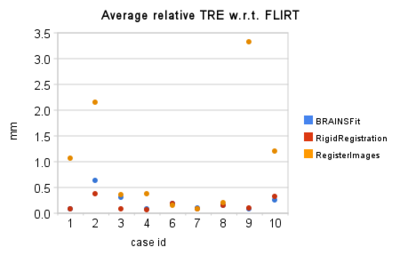
- quantitative assessment: relative Target Registration Error (TRE) statistics collected per-pixel over the segmented tumor volume. In the absence of ground truth for registration result, and considering the wide acceptance and thorough testing of FSL FLIRT registration tool, we accept its result to be optimal under existing operating conditions. We then evaluate each of the registration tools as follows:
- Let T_{flirt} and T_{reg} be the transforms aligning the images being registered recovered with FSL FLIRT and the compared registration tool, respectively
- Let T(p) be the coordinates of a point after applying a transform
- We then define the relative registration error e as follows: e = ||T_{flirt}(p)-T_{reg}(p)||
- Given the binary mask defining the tumor, we evaluate the value of registration error surrogate at each pixel corresponding to the tumor volume, and report the values of min, max, and average relative error values for this region.
- minimum execution time to meet the application requirements: interactive quantification of tumor growth
- minimum tuning of registration parameters (is this possible? to answer this sub-question we compare the results with FLIRT registration using its default parameters)
Methods
Registration
RegisterImages Slicer module
- parameters:
Slicer3 --launch RegisterImages --saveTransform ${RI_TFM} \
--initialization ImageCenters --registration Rigid --expectedOffset 10 \
--expectedRotation 0.1 --expectedScale 0.05 --expectedSkew 0.01 \
--verbosityLevel Standard --sampleIntensityPortion 0.05 --randomNumberSeed 12345 \
--numberOfThreads 4 --baselineNumberOfFailedPixelsTolerance 1000 \
--baselineIntensityTolerance 10 --baselineRadiusTolerance 0 --rigidMaxIterations 200 \
--rigidSamplingRatio 0.01 --affineMaxIterations 50 --affineSamplingRatio 0.02 \
--bsplineMaxIterations 20 --bsplineSamplingRatio 0.1 --controlPointSpacing 40 \
${FIXED_IMAGE} ${MOVING_IMAGE}
- execution time: ~1-2 min
RigidRegistration Slicer module
- parameters
Slicer3 --launch RigidRegistration --fixedsmoothingfactor 0 \
--movingsmoothingfactor 0 --histogrambins 50 --spatialsamples 100000 \
--iterations 1000,1000,500,200 --learningrate 0.01,0.005,0.0005,0.0002 \
--translationscale 100 --outputtransform ${RR_TFM}
${FIXED_IMAGE} ${MOVING_IMAGE}
NOTE: the default value for the number of samples was increased by 10, and the number of histogram bins was increased to 50 from 30.
- execution time: ~4min
BRAINSFit
- BRAINSFit on NITRC
- Technical details on InsightJournal
- Parameters:
${SLICER} --launch BRAINSFitExe \
--fixedVolume ${TIME_POINT_1} --movingVolume ${TIME_POINT_2} \
--transformType Rigid --initializeTransformMode CenterOfHead \
--numberOfIterations 200 --numberOfSamples 100000 --minimumStepSize 0.005 \
--spatialScale 1000 --reproportionScale 1 \
--outputTransform ${BF_TRANSFORM} --histogramMatch \
--strippedOutputTransform ${BF_STRIPPED_TRANSFORM} \
--patientID ANON --studyID ANON --outputVolumePixelType float \
--backgroundFillValue 0 --maskProcessingMode ROIAUTO \
--fixedVolumeTimeIndex 0 --movingVolumeTimeIndex 0 \
--fixedVolumeOrigin 0,0,0 --movingVolumeOrigin 0,0,0 \
--medianFilterSize 0,0,0 --permitParameterVariation 1,1,1 \
--useCachingOfBSplineWeightsMode ON --useExplicitPDFDerivativesMode Whatever \
--relaxationFactor 0.5 --maximumStepSize 0.2 --failureExitCode -1 \
--fixedVolumeROI ${FIXED_IMAGE_ROI} --movingVolumeROI ${MOVING_IMAGE_ROI} \
--numberOfHistogramBins 50 --numberOfMatchPoints 10 --debugNumberOfThreads 4
Note: all parameters are the default settings except numberOfIterations (default 1500), maskProcessingMode (default NOMASK).
- execution time: ~12 min. Breakdown for Case01: CenterOfHead transform initialization: 343s, ROIAUTO mask detection: 139s + 181s (fixed + moving), Rigid registration: 46s
FSL FLIRT
- FSL FLIRT
- Technical details
- parameters:
flirt -in ${TIME_POINT_2} -ref ${TIME_POINT_1} -omat ${FLIRT_TRANSFORM} -out ${RESAMPLED_IMAGE} -dof 6 -v
- execution time: ~4 min
Parameter exploration
Validation and accuracy assessment
Results
Qualitative assessment
BRAINSFit
A distinctive feature of BRAINSFit tool is the ability to automatically detect head region of interest, which is used for collecting intensity samples during metric calculation. Below is the overlay of such ROI in light blue overlayed with the image for Case10. This functionality appears to work consistently for all the cases used in the comparison.
- Case01
- Case03
- Case04
- Case10
FSL FLIRT
- Case01
- Case03
- Case04
- Case10
Quantitative comparison
Execution times recap (UPDATE: these times are for the code compiled in debug mode! In the release mode, execution time of BRAINSFit is under 1 minute):
- FLIRT: ~ 4 min
- BRAINSFit: > 10 min (with most of the time, ~80%, spent in ROI detection and transform initialization)
- RigidRegistration: ~ 4 min
- RegisterImages: ~ 2 min
Conclusions
Limitations of the Slicer registration tools
- RigidRegistration (the following are trivial to fix in the code)
- Number of samples is specified as an absolute value, better if it is a relative value to the total number of the image voxels
- Not possible to specify input mask, or sampling threshold for metric voxels
- Not possible to specify the number of threads, while variable number of threads used in registration may result in irreproducible results (see ITK bug #9222
- RegisterImages
- Output transform should be loaded manually via Transform IO (convenience). Not trivial to fix, since RegisterImages is generic to use the same output transform for different types of registration (e.g., Rigid, Affine and BSpline), while BSpline cannot be loaded in Slicer via transform IO
- all of the tools implemented in Slicer require to specify the max number of iterations, which essentially requires the user to monitor convergence (metric value while iterating can be found in Slicer error log), and increase the number of iterations if the satisfactory result is not achieved. Not practical for interactive application
- not possible to specify the similarity metric: apparently, FLIRT is able to get good result using cross correlation, which is less demanding computationally
- RegisterImages and RigidRegistration do not allow to specify 9-dof transforms, which are reportedly used to adjust for voxel size inconsistency in longitudinal studies [1].
Acknowledgements
We acknowledge and appreciate the efforts of Hans Johnson, Jim Miller and Stephen Aylward in developing open source registration tools analyzed here. FSL FLIRT tool was used here solely for academic research purposes. We thank Luke Bloy for providing the code to perform conversion of FLIRT transform matrix into the ITK-readable format.
References
- J. L. Whitwell, J. M. Schotta, E. B. Lewis, D. G. Macmanus, and N. C. Fox, "Using nine degrees-of-freedom registration to correct for changes in voxel size in serial mri studies," Magnetic Resonance Imaging, vol. 22, no. 7, pp. 993-999, September 2004. [1]