MeningiomaMRIRegistrationStudy
From NAMIC Wiki
Home < MeningiomaMRIRegistrationStudy
Contents
Objective
Accurate registration of same patient/same modality MRI data for longitudinal analysis of tumor progression.
Specific Aims
- Compare the accuracy of registration using existing Slicer and non-Slicer tools
- Identify parameter settings that produce satisfactory results
- Outline the limitations of the available registration tools in the context of the specific clinical research application
Data
- Input images: isotropic post-contrast T1 MRI acquired at different locations of BWH during 2006-2008, used under medical records study IRB. Time period between acquisition of scans for each patient is about 1 year.
- Ground truth transformation: not available
- Expert landmarks for registration evaluation: not available
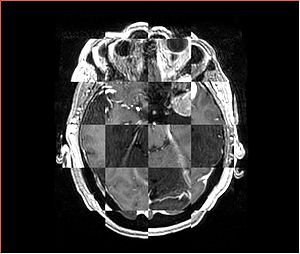
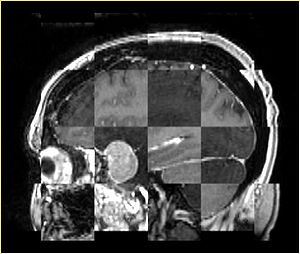
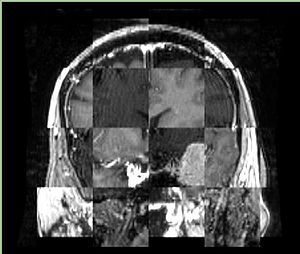
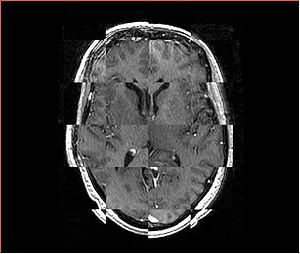
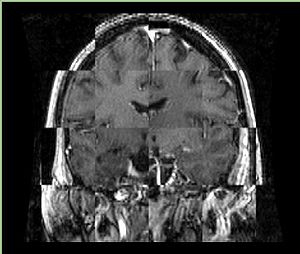
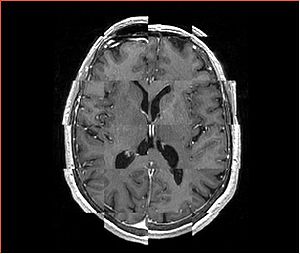
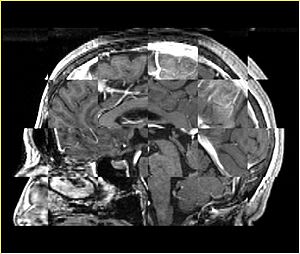
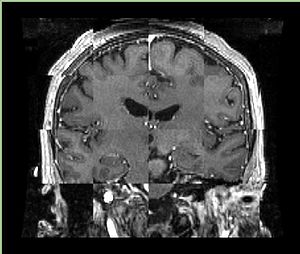
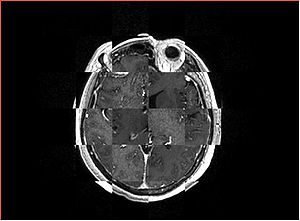
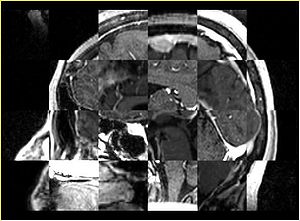
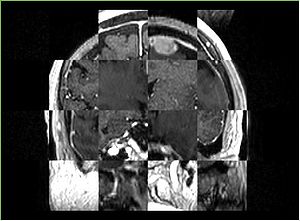
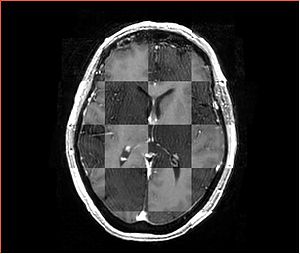
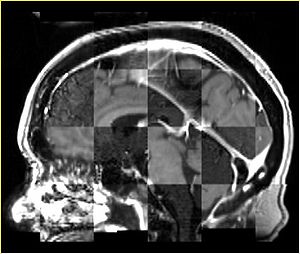
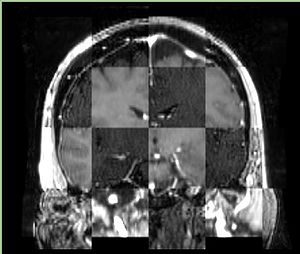
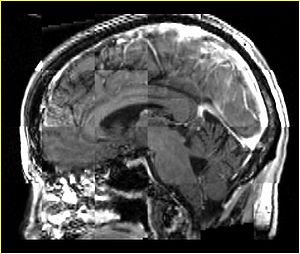
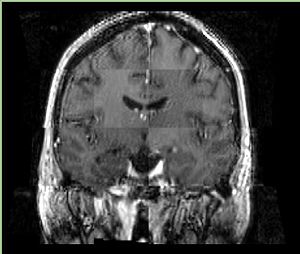
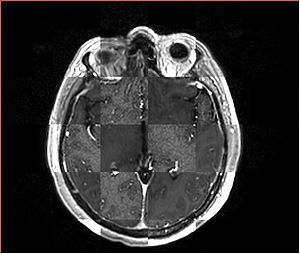
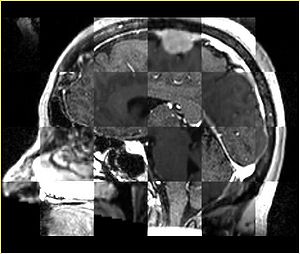
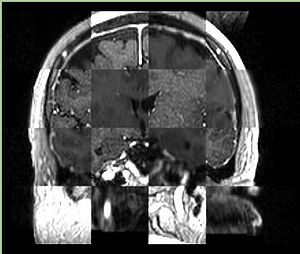
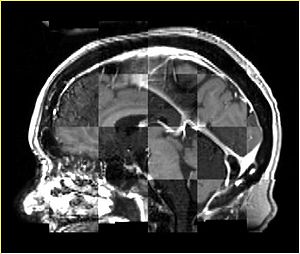
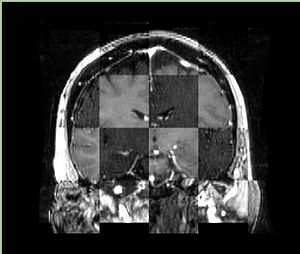
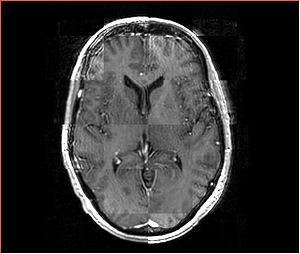
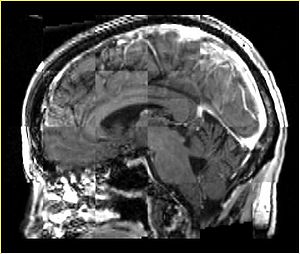
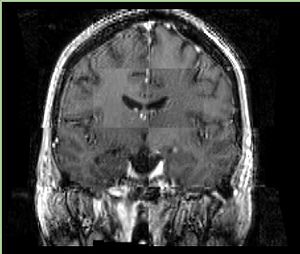
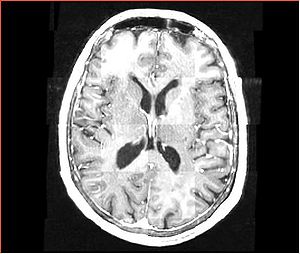
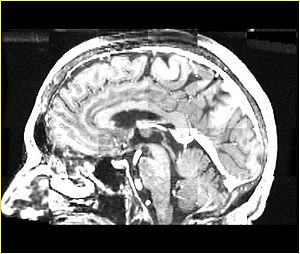
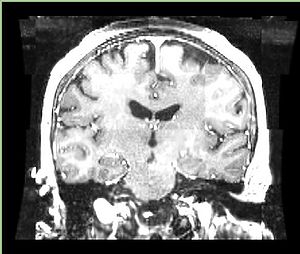
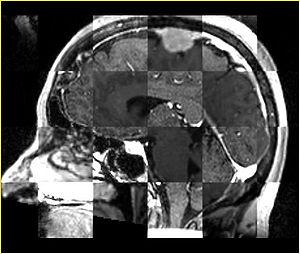
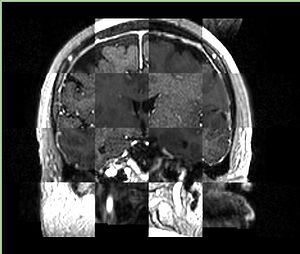
- Checkerboard appearance of unregistered images for the representative data of interest
- Case01
- Case03
- Case04
- Case10
Environment
All registration methods were executed on george.bwh.harvard.edu. Where number of threads was possible to specify, it was set to 4.
Measures of success
- qualitative assessment: visually pleasing results
- quantitative assessment: something better than "visually pleasing" (TBD)
- minimum execution time to meet the application requirements: interactive quantification of tumor growth
- minimum tuning of registration parameters
Methods
Registration
RegisterImages Slicer module
RigidRegistration Slicer module
BRAINSFit
- BRAINSFit on NITRC
- Technical details on InsightJournal
- Parameters:
${SLICER} --launch BRAINSFitExe \
--fixedVolume ${TIME_POINT_1} --movingVolume ${TIME_POINT_2} \
--transformType Rigid --initializeTransformMode CenterOfHead \
--numberOfIterations 200 --numberOfSamples 100000 --minimumStepSize 0.005 \
--spatialScale 1000 --reproportionScale 1 \
--outputTransform ${BF_TRANSFORM} --histogramMatch \
--strippedOutputTransform ${BF_STRIPPED_TRANSFORM} \
--patientID ANON --studyID ANON --outputVolumePixelType float \
--backgroundFillValue 0 --maskProcessingMode ROIAUTO \
--fixedVolumeTimeIndex 0 --movingVolumeTimeIndex 0 \
--fixedVolumeOrigin 0,0,0 --movingVolumeOrigin 0,0,0 \
--medianFilterSize 0,0,0 --permitParameterVariation 1,1,1 \
--useCachingOfBSplineWeightsMode ON --useExplicitPDFDerivativesMode Whatever \
--relaxationFactor 0.5 --maximumStepSize 0.2 --failureExitCode -1 \
--fixedVolumeROI ${FIXED_IMAGE_ROI} --movingVolumeROI ${MOVING_IMAGE_ROI} \
--numberOfHistogramBins 50 --numberOfMatchPoints 10 --debugNumberOfThreads 4
Note: all parameters are the default settings except numberOfIterations (default 1500), maskProcessingMode (default NOMASK).
- execution time: ~12 min
FSL FLIRT
- FSL FLIRT
- Technical details
- parameters:
flirt -in ${TIME_POINT_2} -ref ${TIME_POINT_1} -omat ${FLIRT_TRANSFORM} -out ${RESAMPLED_IMAGE} -dof 6 -v
- execution time: ~4 min
Parameter exploration
Validation and accuracy assessment
Results
BRAINSFit
- Case01
- Case03
- Case04
- Case10
FSL FLIRT
- Case01
- Case03
- Case04
- Case10