|
|
| (One intermediate revision by the same user not shown) |
| Line 1: |
Line 1: |
| | {| | | {| |
| | |[[Image:ProjectWeek-2008.png|thumb|320px|Return to [[2008_Summer_Project_Week|Project Week Main Page]] ]] | | |[[Image:ProjectWeek-2008.png|thumb|320px|Return to [[2008_Summer_Project_Week|Project Week Main Page]] ]] |
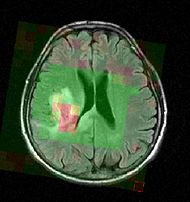
| − | |[[Image:genuFAp.jpg|thumb|320px|Scatter plot of the original FA data through the genu of the corpus callosum of a normal brain.]] | + | |[[Image:Mrsi slicer.jpg|thumb|190px|Color-coded MRSI in the localization and grading of brain tumors.]] |
| − | |[[Image:genuFA.jpg|thumb|320px|Regression of FA data; solid line represents the mean and dotted lines the standard deviation.]]
| |
| | |} | | |} |
| − |
| |
| − |
| |
| | __NOTOC__ | | __NOTOC__ |
| − |
| |
| − | ===Instructions for Use of this Template===
| |
| − | #Please create a new wiki page with an appropriate title for your project using the convention NA-MIC/Projects/Theme-Name/Project-Name
| |
| − | #Copy the entire text of this page into the page created above
| |
| − | #Link the created page into the list of projects for the project event
| |
| − | #Delete this section from the created page
| |
| − | #Send an email to tkapur at bwh.harvard.edu if you are stuck
| |
| − |
| |
| − | ===Key Investigators===
| |
| − | * UNC: Isabelle Corouge, Casey Goodlett, Guido Gerig
| |
| − | * Utah: Tom Fletcher, Ross Whitaker
| |
| − |
| |
| − |
| |
| | <div style="margin: 20px;"> | | <div style="margin: 20px;"> |
| − | | + | <div style="width: 40%; float: left; padding-right: 3%;"> |
| − | <div style="width: 27%; float: left; padding-right: 3%;"> | |
| − | | |
| | <h1>Objective</h1> | | <h1>Objective</h1> |
| − | We are developing methods for analyzing diffusion tensor data along fiber tracts. The goal is to be able to make statistical group comparisons with fiber tracts as a common reference frame for comparison.
| + | Magnetic resonance spectroscopic imaging (MRSI) is a non-invasive diagnostic method used to determine the relative abundance of specific metabolites at arbitrary locations in vivo. Certain diseases - such as tumors in brain, breast and prostate - can be can be associated with characteristic changes in the metabolic level. Thus, proton MRSI is in principle very well suited for the detection, localization and grading of these diseases. A major challenge in MRSI, however, lies in the postprocessing and evaluation of the acquired spectral volumes, and the availability of MRSI data processing routines. |
| − | | |
| | | | |
| | + | The objective of the current project is to develop a module proving the means for the processing and visualization of MRSI, and thus for a joint analysis of magnetic resonance spectroscopic images together with other imaging modalities in Slicer. |
| | </div> | | </div> |
| | | | |
| | <div style="width: 27%; float: left; padding-right: 3%;"> | | <div style="width: 27%; float: left; padding-right: 3%;"> |
| − |
| |
| | <h1>Approach, Plan</h1> | | <h1>Approach, Plan</h1> |
| − | | + | The standard approach in the analysis of magnetic resonance spectra is the fitting of resonance-line shaped model functions to the spectral pattern, often referred to as "quantification". The most likely parameter estimate for a given signal model is determined with a nonlinear least squares approach, additional steps in the signal processing remove broad baselines and residuals of the water peak. |
| − | Our approach for analyzing diffusion tensors is summarized in the IPMI 2007 reference below. The main challenge to this approach is <foo>.
| |
| − | | |
| − | Our plan for the project week is to first try out <bar>,...
| |
| | </div> | | </div> |
| | | | |
| − | <div style="width: 40%; float: left;"> | + | <div style="width: 27%; float: left;"> |
| − | | |
| | <h1>Progress</h1> | | <h1>Progress</h1> |
| − | | + | Defined a signal processing pipeline and identified resources for the implementation. |
| − | Software for the fiber tracking and statistical analysis along the tracts has been implemented. The statistical methods for diffusion tensors are implemented as ITK code as part of the [[NA-MIC/Projects/Diffusion_Image_Analysis/DTI_Software_and_Algorithm_Infrastructure|DTI Software Infrastructure]] project. The methods have been validated on a repeated scan of a healthy individual. This work has been published as a conference paper (MICCAI 2005) and a journal version (MEDIA 2006). Our recent IPMI 2007 paper includes a nonparametric regression method for analyzing data along a fiber tract.
| |
| | | | |
| | </div> | | </div> |
| | | | |
| | <br style="clear: both;" /> | | <br style="clear: both;" /> |
| − |
| |
| | </div> | | </div> |
| − | | + | ===Key Investigators=== |
| − | | + | * SPL: Bjoern Menze |
| − | ===References===
| |
| − | * Fletcher, P.T., Tao, R., Jeong, W.-K., Whitaker, R.T., "A Volumetric Approach to Quantifying Region-to-Region White Matter Connectivity in Diffusion Tensor MRI," to appear Information Processing in Medical Imaging (IPMI) 2007.
| |
| − | * Corouge, I., Fletcher, P.T., Joshi, S., Gilmore, J.H., and Gerig, G., "Fiber Tract-Oriented Statistics for Quantitative Diffusion Tensor MRI Analysis," Medical Image Analysis 10 (2006), 786--798.
| |
| − | * Corouge, I., Fletcher, P.T., Joshi, S., Gilmore J.H., and Gerig, G., Fiber Tract-Oriented Statistics for Quantitative Diffusion Tensor MRI Analysis, Lecture Notes in Computer Science LNCS, James S. Duncan and Guido Gerig, editors, Springer Verlag, Vol. 3749, Oct. 2005, pp. 131 -- 138
| |
| − | * C. Goodlett, I. Corouge, M. Jomier, and G. Gerig, A Quantitative DTI Fiber Tract Analysis Suite, The Insight Journal, vol. ISC/NAMIC/ MICCAI Workshop on Open-Source Software, 2005, Online publication: http://hdl.handle.net/1926/39
| |
| − | ===Text from Adam's Previous Page===
| |
| − | Back to [[NA-MIC_Collaborations|NA-MIC_Collaborations]], [[Algorithm:Site1|Site1 Algorithms]], [[DBP1:Site2|Site2 DBP 1]]
| |
| − | __NOTOC__
| |
| − | = '''Computing the Brain Deformation for Image-Guided Neurosurgery''' (The Intelligent Systems of Medicine Laboratory [http://www.mech.uwa.edu.au/ISML/ link title] | |
| − | School of Mechanical Engineering, The University of Western Australia)=
| |
| − | | |
| − | =Description= | |
| − | | |
| − | The Intelligent Systems of Medicine Laboratory (ISML) mission is to work towards improving clinical outcomes through appropriate use of technology. We are interested in biomechanics (both engineering biomechanics and sport biomechanics), biomedical engineering, computer integrated surgery, medical robotics and related fields. We run exciting research projects in these areas, generously funded by The Australian Research Council and other agencies.
| |
| − | | |
| − | We intend to contribute to Na-MIC by providing algorithms for computing the intra-operative brain deformations for image-guided neurosurgery. We treat the brain shift as a continuum mechanics problem involving finite deformations and solve it using non-linear finite element procedures. We use the procedures (non-linear explicit dynamics with Total Lagrangian formulation) that do not require iterations even when applied to non-linear problems and are, therefore, amenable to computing the intra-operative brain deformations in real time (under 150 s) on a standard PC.
| |
| − | | |
| − | | |
| − | = Key Investigators = | |
| − | | |
| − | * Prof. Karol Miller (kmiller@mech.uwa.edu.au), Dr Adam Wittek, Grand Joldes
| |
| − | | |
| − | = Publications [http://www.mech.uwa.edu.au/ISML/publications.htm link title]=
| |
| − | | |
| − | *[1] Miller, K., Joldes, G., Lance, D., Wittek, A. (2007) Total Lagrangian explicit dynamics finite element algorithm for computing soft tissue deformation, Communications in Numerical Methods in Engineering. Vol. 23, pp. 121-134, doi: 10.1002/cnm.887, available on-line at [http://www3.interscience.wiley.com/cgi-bin/jissue/109075715 link title].
| |
| − | *[2] Wittek, A., Miller, K., Kikinis, R., Warfield, S. K. (2007) Patient-specific model of brain deformation: Application to medical image registration. Journal of Biomechanics. Vol. 40, pp. 919-929, DOI:10.1016/j.jbiomech.2006.02.021, available on-line at [http://www.sciencedirect.com/ link title].
| |
| − | *[3] Joldes, G. R., Wittek, A., Miller, K. (2007) Suite of finite element algorithms for accurate computation of soft tissue deformation for surgical simulation, in Proceedings of Computational Biomechanics for Medicine Workshop, International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2007, Brisbane, Australia, ISBN 13: 978 0 643 09517 5, pp. 65-73.
| |
| − | *[4] Hawkins, T., Wittek, A. and Miller, K. (2006) Comparison of constitutive models of brain tissue for non-rigid image registration, in CD Proceedings of 2nd Workshop on Computer Assisted Diagnosis and Surgery, Santiago, Chile, 4 pages.
| |
| − | *[5] Horton, A., Wittek, A. and K. Miller (2006) Computer simulation of brain shift using an element free Galerkin method, in CD Proceedings of 7th International Symposium on Computer Methods in Biomechanics and Biomedical Engineering CMBBE 2006, Antibes, France, ISBN: 0-9549670-2-X, pp. 906-911.
| |
| − | *[6] Horton, A., Wittek, A. and K. Miller (2006) Towards meshless methods for surgery simulation. Linear versus non-linear computation of the brain shift, in Proceedings of Computational Biomechanics for Medicine Workshop, International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2006, Copenhagen, Denmark, ISBN 10: 87-7611-149-0, pp. 32-40 [http://www.mech.uwa.edu.au/ISML/publications.htm link title].
| |
| − | *[7] Joldes, G., Wittek, A. and Miller, K. (2006) Improved linear tetrahedral element for surgery simulation, in Proceedings of Computational Biomechanics for Medicine Workshop, International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2006, Copenhagen, Denmark, ISBN 10: 87-7611-149-0, pp. 52-63 [http://www.mech.uwa.edu.au/ISML/ publications.htm link title].
| |
| − | *[8] Joldes, G., Wittek, A. and Miller, K. (2006) Towards non-linear finite element, computations in real time, in CD Proceedings of 7th International Symposium on Computer Methods in Biomechanics and Biomedical Engineering CMBBE 2006, Antibes, France, ISBN: 0-9549670-2-X, pp. 894-899.
| |
| − | *[9] Miller, K., Joldes, G. and Wittek, A. (2006) New finite element algorithm for surgical simulation, in CD Proceedings of 2nd Workshop on Computer Assisted Diagnosis and Surgery, Santiago, Chile, 4 pages
| |
| − | *[10] Miller, K. Hawkins, T. and Wittek, A. (2006) Linear versus non-linear computation of the brain shift, in CD Proceedings of the 7th International Symposium on Computer Methods in Biomechanics and Biomedical Engineering CMBBE 2006, Antibes, France, ISBN: 0-9549670-2-X, pp. 888-893.
| |
| − | *[11] Miller, K. and Wittek, A. (2006) Neuroimage registration as displacement - zero traction problem of solid mechanics, Lead Lecture in Proceedings of Computational Biomechanics for Medicine Workshop, International Conference on Medical Image Computing and Computer-Assisted Intervention MICCAI 2006, Copenhagen, Denmark, ISBN 10: 87-7611-149-0, pp. 1-12 [http://www.mech.uwa.edu.au/ISML/ publications.htm link title].
| |
| − | *[12] Wittek, A., Kikinis, K., Warfield, S. K., and Miller, K. (2005) Brain shift computation using a fully nonlinear biomechanical model, in Proceedings of 8th International Conference on Medical Image Computing and Computer Assisted Intervention MICCAI 2005 in Lecture Notes in Computer Science 3750 2006, pp. 583-590.
| |
| − | | |
| − | | |
| − | ''In Press''
| |
| − | *[13] AWittek, A., T. Hawkins, and Miller, K. (2008). On the unimportance of constitutive models in computing brain deformation for image-guided surgery (in press). Biomechanics and Modeling in Mechanobiology: 8 pages, doi: 10.1007/s10237-008-0118-1, Springer.
| |
| − | *[14] Grand Joldes, Karol Miller and Adam Wittek (2007) Efficient hourglass control implementation for an uniform strain hexahedra using Total Lagrangian formulation. Communications in Numerical Methods in Engineering (accepted in June 2007), 9 pages, doi: 10.1002/cnm.1034, Wiley.
| |
| − | | |
| − | = Links = | |
| − | | |
| − | * Link1
| |
| − | * Link2 | |
| − | | |
| − | Project Week Results: [[blah|Jan 2006]], [[blah|Jun 2007]]
| |