Difference between revisions of "NA-MIC/Projects/Collaboration/MRSI Module for Slicer"
| Line 19: | Line 19: | ||
<div style="width: 27%; float: left;"> | <div style="width: 27%; float: left;"> | ||
<h1>Progress</h1> | <h1>Progress</h1> | ||
| + | Defined a signal processing pipeline and identified resources for the implementation. | ||
</div> | </div> | ||
Latest revision as of 12:48, 27 June 2008
Home < NA-MIC < Projects < Collaboration < MRSI Module for Slicer Return to Project Week Main Page |
Objective
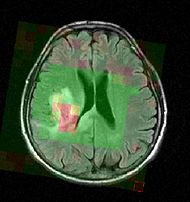
Magnetic resonance spectroscopic imaging (MRSI) is a non-invasive diagnostic method used to determine the relative abundance of specific metabolites at arbitrary locations in vivo. Certain diseases - such as tumors in brain, breast and prostate - can be can be associated with characteristic changes in the metabolic level. Thus, proton MRSI is in principle very well suited for the detection, localization and grading of these diseases. A major challenge in MRSI, however, lies in the postprocessing and evaluation of the acquired spectral volumes, and the availability of MRSI data processing routines.
The objective of the current project is to develop a module proving the means for the processing and visualization of MRSI, and thus for a joint analysis of magnetic resonance spectroscopic images together with other imaging modalities in Slicer.
Approach, Plan
The standard approach in the analysis of magnetic resonance spectra is the fitting of resonance-line shaped model functions to the spectral pattern, often referred to as "quantification". The most likely parameter estimate for a given signal model is determined with a nonlinear least squares approach, additional steps in the signal processing remove broad baselines and residuals of the water peak.
Progress
Defined a signal processing pipeline and identified resources for the implementation.
Key Investigators
- SPL: Bjoern Menze