NA-MIC/Projects/Collaboration/SBIA UPenn
From NAMIC Wiki
Home < NA-MIC < Projects < Collaboration < SBIA UPenn
DTI and HARDI analysis tools
 Return to Project Week Main Page |
Key Investigators
- Ragini Verma, SBIA, UPenn <Ragini.Verma@uphs.upenn.edu>
- Christos Davatzikos <Christos.Davatzikos@uphs.upenn.edu>
- Yang Li <Yang.Li@uphs.upenn.edu> (DTI integration)
- Luke Bloy <lbloy@seas.upenn.edu> (HARDI integration)
- Jinzhong Yang
- Dinggang Shen
- Carl-Fredrik Westin, BWH
- Luis Ibanez, Slicer
- Steve Pieper, Slicer
Objective
To incorporate into Slicer, processing and analysis methods for DTI and HARDI being developed at the Section of Biomedical Image Analysis (SBIA), UPenn [1]. The main components will be plugins for DTI registration and manifold-based statistics followed by methods for HARDI registration and statistics.
Approach, Plan
- DTI registration: has been developed in SBIA over a period of 2 years, mainly using in-house code (not in ITK). This will be incorporated as a plugin.
- DTI Statistics: SPM-like package for statistics on the full tensor. The proposed format is again a plugin as it is based on code already existing in SBIA in C, C++.
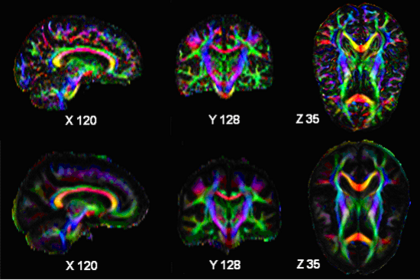
- HARDI registration: This is being developed in ITK, building on HARDI representation format currently under development
- HARDI statistics: being developed in ITK
Progress
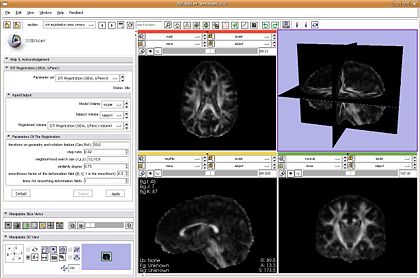
- DTI Registration: plugin is ready and will be demonstrated
To do: Replace the affine registration component, help file, speed, version control
- Need help with:
1. General representation for registration in ITK
2. Code for Demons DTI registration for comparison
3. Visualization: tensor glyphs and ODFs
References
This work was supported in part by NIH grants R01-MH-079938 and R01-MH-070365.
- Link to SBIA web site:
- J. Yang, D. Shen, C. Davatzikos and R. Verma, Diffusion Tensor Image Registration Using Tensor Geometry and Orientation Features, in MICCAI 2008, New York.
- J. Yang, D. Shen, C. Davatzikos and R. Verma, Spatial Normalization of Diffusion Tensor Images Based on Anisotropic Segmentation, in SPIE Medical Imaging 2008, San Diego.
- R. Verma, P. Khurd, C. Davatzikos, On Analyzing Diffusion Tensor Images by Identifying Manifold Structure using Isomaps, IEEE Transactions on Medical Imaging, 772-778, Vol. 26, No. 6, 2007
- P. Khurd, R. Verma and C. Davatzikos, Kernel-based Manifold Learning for Statistical Analysis of Diffusion Tensor Images, Information Processing in Medical Imaging (IPMI), 581-593, Vol. 4584, 2007