Difference between revisions of "NA-MIC/Projects/NA-MIC Kit/Execution Model"
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
|||
| (8 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
Back to [[NA-MIC_Collaborations|NA-MIC_Collaborations]] | Back to [[NA-MIC_Collaborations|NA-MIC_Collaborations]] | ||
| − | + | = Name of Template Project goes here = | |
| − | + | The Slicer3 Execution Model realizes the "Extensible Algorithmic Framework" goal by providing an interactive point-and-click interface to ITK-based command line executables as well as a batch processing interface to execute large scale experiments on clusters and grids. This objective is reached without requiring extensive re-engineering by the algorithm developer. The Execution Model uses an XML syntax to describe algorithm options and user interface controls. Slicer3 constructs the appropriate user interfaces from this description and manages the data transfer to and from the algorithm module. Modules are discovered at runtime and delivered to clinicians as plugins to Slicer3. | |
| + | = Description = | ||
| − | + | A base implementation of module discovery, user interface construction, and module execution is integrated into Slicer3. The implementation supports modules packaged as command line executables as well as modules loaded as dynamic libraries. Modules can provide their own help information, acknowledgments, and logos. A rich set of parameter types can be provided to a module including: scalar images, vector images, diffusion weighted images, diffusion tensor images, geometry, points, booleans, integers, floating point numbers, strings, files, directories, lists, and enumerations. Additional parameter datatypes such as MRML scenes, comma separated value files, and transforms are under development. Parameter sets can be archived as part of a MRML scene in Slicer3. Parameters are realized in the Slicer3 user interface using a rich set of widgets that include: node selectors, sliders, text entries, spin boxes, check boxes, radio buttons, and directory browsers. Modules can report multistage progress and modules can be terminated from Slicer3. Modules are run in a separate thread of execution and are queued so that only one module will execute at a time. Modules report information to the standard Slicer3 logging facility. Extensive documentation on the execution model is available describing the XML module description, the parameter types, command line parameter parsing tools, the automatic user interface construction, process control tools, and CMake configurations for new modules. | |
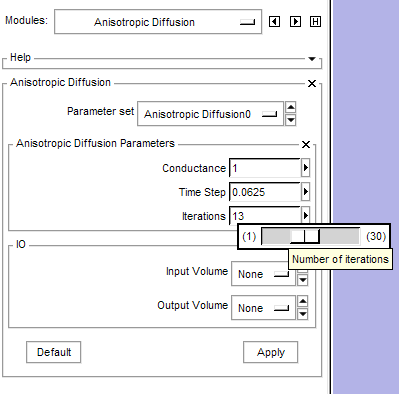
| − | + | [[Image:AnisotropicDiffusionFilterGUI.png|thumb|400px|Slicer3 user interface for a command line module. This user interface was constructed automatically from the XML description of the module parameters.]] | |
| − | |||
| − | * [ | + | = Key Investigators = |
| − | * [ | + | |
| + | *GE: Dan Blezek,Jim Miller and Bill Lorensen | ||
| + | |||
| + | = Links = | ||
| + | |||
| + | * [https://www.slicer.org/wiki/Slicer3:Execution_Model Slicer3 Execution Model] | ||
| + | * [https://www.slicer.org/wiki/Slicer3:Execution_Model_Documentation Slicer3 Execution Model Documentation] | ||
| + | * [https://www.slicer.org/wiki/Slicer3:VisualBlog Slicer3 Visual Blog] | ||
Latest revision as of 17:09, 10 July 2017
Home < NA-MIC < Projects < NA-MIC Kit < Execution ModelBack to NA-MIC_Collaborations
Name of Template Project goes here
The Slicer3 Execution Model realizes the "Extensible Algorithmic Framework" goal by providing an interactive point-and-click interface to ITK-based command line executables as well as a batch processing interface to execute large scale experiments on clusters and grids. This objective is reached without requiring extensive re-engineering by the algorithm developer. The Execution Model uses an XML syntax to describe algorithm options and user interface controls. Slicer3 constructs the appropriate user interfaces from this description and manages the data transfer to and from the algorithm module. Modules are discovered at runtime and delivered to clinicians as plugins to Slicer3.
Description
A base implementation of module discovery, user interface construction, and module execution is integrated into Slicer3. The implementation supports modules packaged as command line executables as well as modules loaded as dynamic libraries. Modules can provide their own help information, acknowledgments, and logos. A rich set of parameter types can be provided to a module including: scalar images, vector images, diffusion weighted images, diffusion tensor images, geometry, points, booleans, integers, floating point numbers, strings, files, directories, lists, and enumerations. Additional parameter datatypes such as MRML scenes, comma separated value files, and transforms are under development. Parameter sets can be archived as part of a MRML scene in Slicer3. Parameters are realized in the Slicer3 user interface using a rich set of widgets that include: node selectors, sliders, text entries, spin boxes, check boxes, radio buttons, and directory browsers. Modules can report multistage progress and modules can be terminated from Slicer3. Modules are run in a separate thread of execution and are queued so that only one module will execute at a time. Modules report information to the standard Slicer3 logging facility. Extensive documentation on the execution model is available describing the XML module description, the parameter types, command line parameter parsing tools, the automatic user interface construction, process control tools, and CMake configurations for new modules.
Key Investigators
- GE: Dan Blezek,Jim Miller and Bill Lorensen