|
|
| (20 intermediate revisions by one other user not shown) |
| Line 1: |
Line 1: |
| | Back to [[NA-MIC_Collaborations:New|NA-MIC Collaborations]] | | Back to [[NA-MIC_Collaborations:New|NA-MIC Collaborations]] |
| | __NOTOC__ | | __NOTOC__ |
| − | == Diffusion Image Analysis == | + | = NA-MIC Internal Collaborations = |
| | | | |
| − | === Fiber Tract Extraction and Analysis === | + | {| cellpadding="10" border="1" |
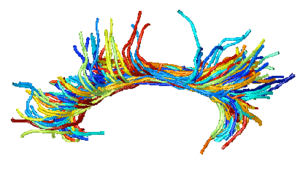
| | + | | style="width:33%" | [[Image:CingulumAllSubjectsFibers.png|300px]] |
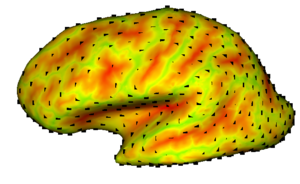
| | + | | style="width:33%" | [[Image:Sulcaldepth.png|300px]] |
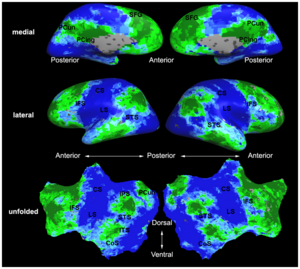
| | + | | style="width:33%" | [[Image:Mit_fmri_clustering_parcellation2_xsub.png|300px]] |
| | | | |
| − | {| cellpadding="10"
| + | |-align="center" |
| − | | style="width:15%" | [[Image:ZoomedResultWithModel.png|200px]]
| |
| − | | style="width:85%" |
| |
| − | | |
| − | == [[Projects:GeodesicTractographySegmentation|Geodesic Tractography Segmentation]] ==
| |
| − | | |
| − | In this work, we provide an energy minimization framework which allows one to find fiber tracts and volumetric fiber bundles in brain diffusion-weighted MRI (DW-MRI). [[Projects:GeodesicTractographySegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> J. Melonakos, E. Pichon, S. Angenet, and A. Tannenbaum. Finsler Active Contours. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2007.
| |
| − | | |
| − | |}
| |
| − | | |
| − | === Fractional Anisotropy Analysis ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" |
| |
| − | | style="width:85%" |
| |
| − | | |
| − | |}
| |
| − | | |
| − | === Path of Interest Analysis ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" |
| |
| − | | style="width:85%" |
| |
| − | | |
| − | |}
| |
| − | | |
| − | === Validation ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" |
| |
| − | | style="width:85%" |
| |
| − | | |
| − | |}
| |
| − | | |
| − | === Algorithm/Software Infrastructure ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" |
| |
| − | | style="width:85%" |
| |
| − | | |
| − | |}
| |
| − | | |
| − | == Structural Image Analysis ==
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" |
| |
| − | | style="width:85%" |
| |
| − | | |
| − | |}
| |
| − | | |
| − | === Image Segmentation ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" | [[Image:Fig67.png|200px|]]
| |
| − | | style="width:85%" |
| |
| − | | |
| − | == [[Projects:KnowledgeBasedBayesianSegmentation|Knowledge-Based Bayesian Segmentation]] ==
| |
| − | | |
| − | This ITK filter is a segmentation algorithm that utilizes Bayes's Rule along with an affine-invariant anisotropic smoothing filter. [[Projects:KnowledgeBasedBayesianSegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> J. Melonakos, Y. Gao, and A. Tannenbaum. Tissue Tracking: Applications for Brain MRI Classification. SPIE Medical Imaging, 2007.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:Striatum1.png|200px|]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:RuleBasedStriatumSegmentation|Rule-Based Striatum Segmentation]] ==
| |
| − | | |
| − | In this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the striatum. [[Projects:RuleBasedStriatumSegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> Al-Hakim, et al. Parcellation of the Striatum. SPIE MI 2007.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:Dlpfc1.jpg|200px|]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:RuleBasedDLPFCSegmentation|Rule-Based DLPFC Segmentation]] ==
| |
| − | | |
| − | In this work, we provide software to semi-automate the implementation of segmentation procedures based on expert neuroanatomist rules for the dorsolateral prefrontal cortex. [[Projects:RuleBasedDLPFCSegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> Al-Hakim, et al. A Dorsolateral Prefrontal Cortex Semi-Automatic Segmenter. SPIE MI 2006.
| |
| − | | |
| − | |-
| |
| − | | |
| − | | | [[Image:Gatech caudateBands.PNG|200px]]
| |
| − | | |
| |
| − | | |
| − | == [[Projects:MultiscaleShapeSegmentation|Multiscale Shape Segmentation Techniques]] ==
| |
| − | | |
| − | To represent multiscale variations in a shape population in order to drive the segmentation of deep brain structures, such as the caudate nucleus or the hippocampus. [[Projects:MultiscaleShapeSegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> Delphine Nain won the best student paper at [[MICCAI_2006|MICCAI 2006]] in the category "Segmentation and Registration" for her paper entitled "Shape-driven surface segmentation using spherical wavelets" by D. Nain, S. Haker, A. Bobick, A. Tannenbaum.
| |
| − | | |
| − | |-
| |
| | | | |
| − | | | [[Image:Stochastic-snake.png|200px|]]
| |
| | | | | | | | |
| | + | == [[NA-MIC_Internal_Collaborations:DiffusionImageAnalysis|Diffusion Image Analysis]] == |
| | | | |
| − | == [[Projects:StochasticMethodsSegmentation|Stochastic Methods for Segmentation]] ==
| |
| − |
| |
| − | New stochastic methods for implementing curvature driven flows for various medical tasks such as segmentation. [[Projects:StochasticMethodsSegmentation|More...]]
| |
| − |
| |
| − | <font color="red">'''New: '''</font> Currently under investigation.
| |
| − |
| |
| − | |-
| |
| − |
| |
| − | | | [[Image:Gatech SlicerModel2.jpg|200px]]
| |
| | | | | | | | |
| | + | == [[NA-MIC_Internal_Collaborations:StructuralImageAnalysis|Structural Image Analysis]] == |
| | | | |
| − | == [[Projects:StatisticalSegmentationSlicer2|Statistical/PDE Methods using Fast Marching for Segmentation]] ==
| |
| − |
| |
| − | This Fast Marching based flow was added to Slicer 2. [[Projects:StatisticalSegmentationSlicer2|More...]]
| |
| − |
| |
| − | |-
| |
| − |
| |
| − | | | [[Image:histo_matching.jpg|200px]]
| |
| | | | | | | | |
| − | | + | == [[NA-MIC_Internal_Collaborations:fMRIAnalysis|fMRI Analysis]] == |
| − | == [[Projects:AutomaticFullBrainSegmentation|Atlas Renormalization for Improved Brain MR Image Segmentation across Scanner Platforms]] == | |
| − | | |
| − | Atlas-based approaches have demonstrated the ability to automatically identify detailed brain structures from 3-D magnetic resonance (MR) brain images. Unfortunately, the accuracy of this type of method often degrades when processing data acquired on a different scanner platform or pulse sequence than the data used for the atlas training. In this paper, we improve the performance of an atlas-based whole brain segmentation method by introducing an intensity renormalization procedure that automatically adjusts the prior atlas intensity model to new input data. Validation using manually labeled test datasets has shown that the new procedure improves the segmentation accuracy (as measured by the Dice coefficient) by 10% or more for several structures including hippocampus, amygdala, caudate, and pallidum. The results verify that this new procedure reduces the sensitivity of the whole brain segmentation method to changes in scanner platforms and improves its accuracy and robustness, which can thus facilitate multicenter or multisite neuroanatomical imaging studies. [[Projects:AutomaticFullBrainSegmentation|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 4, APRIL 2007
| |
| | | | |
| | |} | | |} |
| | | | |
| − | === Image Registration === | + | = Driving Biological Problems (2007 - 2010): Roadmap Projects = |
| | | | |
| − | {| cellpadding="10" | + | {| cellpadding="10" border="1" |
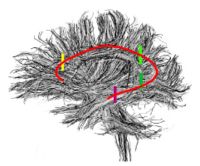
| − | | style="width:15%" | [[Image:Results brain sag.JPG|200px|]] | + | | style="width:15%" | [[Image:Cingulum1.jpg|200px]] |
| | | style="width:85%" | | | | style="width:85%" | |
| | | | |
| − | == [[Projects:OptimalMassTransportRegistration|Optimal Mass Transport Registration]] == | + | == [[ProjectWeek200706:ContrastingTractographyMeasures|Contrasting Tractography Measures]] == |
| | | | |
| − | The goal of this project is to implement a computationaly efficient Elastic/Non-rigid Registration algorithm based on the Monge-Kantorovich theory of optimal mass transport for 3D Medical Imagery. Our technique is based on Multigrid and Multiresolution techniques. This method is particularly useful because it is parameter free and utilizes all of the grayscale data in the image pairs in a symmetric fashion and no landmarks need to be specified for correspondence. [[Projects:OptimalMassTransportRegistration|More...]]
| + | This project represents a new initiative to build upon a shared vision among Cores 1, 3 and 5 that the field of medical image analysis would be well served by work in the area of validation, calibration and assessment of reliability in DW-MRI image analysis. [[ProjectWeek200706:ContrastingTractographyMeasures|More...]] |
| | | | |
| − | <font color="red">'''New: '''</font> Tauseef ur Rehman, A. Tannenbaum. Multigrid Optimal Mass Transport for Image Registration and Morphing. SPIE Conference on Computational Imaging V, Jan 2007. | + | <font color="red">'''New: '''</font> [[SanteFe.Tractography.Conference|Contrasting Tractography Methods Conference]], Santa Fe, October 1-2, 2007. |
| − | | |
| − | |}
| |
| − | | |
| − | === Morphometric Measures and Shape Analysis ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" | [[Image:Basis membership.png|200px]]
| |
| − | | style="width:85%" |
| |
| − | | |
| − | == [[Projects:MultiscaleShapeAnalysis|Multiscale Shape Analysis]] ==
| |
| − | | |
| − | We present a novel method of statistical surface-based morphometry based on the use of non-parametric permutation tests and a spherical wavelet (SWC) shape representation. [[Projects:MultiscaleShapeAnalysis|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> D. Nain, M. Styner, M. Niethammer, J. J. Levitt, M E Shenton, G Gerig, A. Bobick, A. Tannenbaum. Statistical Shape Analysis of Brain Structures using Spherical Wavelets. Accepted in The Fourth IEEE International Symposium on Biomedical Imaging (ISBI ’07) that will be held April 12-15, 2007 in Metro Washington DC, USA.
| |
| | | | |
| | |- | | |- |
| | | | |
| − | | | [[Image:overcomplete_vs_biorthogonal_wavelets.jpg|200px]] | + | | | [[Image:ProstateDiagram.png|200px]] |
| | | | | | | | |
| | | | |
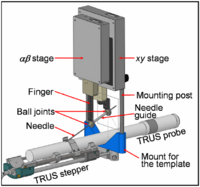
| − | == [[Projects:CorticalSurfaceShapeAnalysisUsingSphericalWavelets|Spherical Wavelets]] == | + | == [[ProjectWeek200706:BrachytherapyNeedlePositioningRobotIntegration|Brachytherapy Needle Positioning Robot Integration]] == |
| − | Cortical Surface Shape Analysis Based on Spherical Wavelets. We introduce the use of over-complete spherical wavelets for shape analysis of 2D closed surfaces. Bi-orthogonal spherical wavelets have been proved to be powerful tools in the segmentation and shape analysis of 2D closed surfaces, but unfortunately they suffer from aliasing problems and are therefore not invariant to rotation of the underlying surface parameterization. In this paper, we demonstrate the theoretical advantage of over-complete wavelets over bi-orthogonal wavelets and illustrate their utility on both synthetic and real data. In particular, we show that the over-complete spherical wavelet transform enjoys significant advantages for the analysis of cortical folding development in a newborn dataset. [[Projects:CorticalSurfaceShapeAnalysisUsingSphericalWavelets|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 4, APRIL 2007
| |
| | | | |
| − | |- | + | The Queen’s/Hopkins team is developing novel devices and procedures for cancer interventions, including biopsy and therapies. Our goal for the programming week is to design and start implementing software for the new MRI Brachytherapy needle positioning robot. [[ProjectWeek200706:BrachytherapyNeedlePositioningRobotIntegration|More...]] |
| | | | |
| − | | | [[Image:separating_loops.jpg|200px]]
| + | <font color="red">'''New: '''</font> Meeting at JHU on July 17-19, 2007. |
| − | | |
| |
| − | | |
| − | == [[Projects:TopologyCorrectionNonSeparatingLoops|Topology Correction]] ==
| |
| − | Geometrically-Accurate Topology-Correction of Cortical Surfaces using Non-Separating Loops. We propose a technique to accurately correct the spherical topology of cortical surfaces. Specifically,we construct a mapping from the original surface onto the sphere to detect topological defects as minimal nonhomeomorphic regions. The topology of each defect is then corrected by opening and sealing the surface along a set of nonseparating loops that are selected in a Bayesian framework. The proposed method is a wholly self-contained topology correction algorithm, which determines geometrically accurate, topologically correct solutions based on the magnetic resonance imaging (MRI) intensity profile and the expected local curvature. Applied to real data, our method provides topological corrections similar to those made by a trained operator. [[Projects:TopologyCorrectionNonSeparatingLoops|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> IEEE TRANSACTIONS ON MEDICAL IMAGING, VOL. 26, NO. 4, APRIL 2007 | |
| | | | |
| | |- | | |- |
| | | | |
| − | | | [[Image:qdec.jpg|200px]] | + | | | [[Image:Lupus.png|200px]] |
| | | | | | | | |
| | | | |
| − | == [[Projects:QDEC|QDEC: An easy to use GUI for group morphometry studies]] == | + | == [[DBP2:MIND:Roadmap|Brain Lesion Analysis in Neuropsychiatric Systemic Lupus Erythematosus]] == |
| | | | |
| − | Qdec is a application included in the Freesurfer software package intended to aid researchers in performing inter-subject / group averaging and inference on the morphometry data (cortical surface and volume) produced by the Freesurfer processing stream. The functionality in Qdec is also available as a processing module within Slicer3, and XNAT. [[Projects:QDEC|More...]]
| + | Our goal is to automatically, or with little or no manual human rater input, accurately tissue classify our example lupus data-set into gray, white, csf, and lesion classes. |
| | | | |
| − | See: [http://surfer.nmr.mgh.harvard.edu/fswiki/Qdec Qdec user page]
| + | <font color="red">'''New: '''</font> Participants in the Summer 2007 NA-MIC Project Week. |
| | | | |
| − | |} | + | |- |
| − | | |
| − | == fMRI Analysis ==
| |
| − | | |
| − | === Functional Activation Analysis ===
| |
| − | | |
| − | {| cellpadding="10"
| |
| − | | style="width:15%" |
| |
| − | | style="width:85%" |
| |
| − | | |
| − | |}
| |
| − | | |
| − | === Algorithm and Software Infrastructure ===
| |
| | | | |
| − | {| cellpadding="10"
| + | | | [[Image:Itksnap.jpg|200px]] |
| − | | style="width:15%" | [[Image:Brain-flat.PNG|200px]]
| + | | | |
| − | | style="width:85%" | | |
| | | | |
| − | == [[Projects:ConformalFlatteningRegistration|Conformal Flattening]] == | + | == [[ProjectWeek200706:CorticalThicknessForAutism|Cortical Thickness for Autism]] == |
| | | | |
| − | The goal of this project is for better visualizing and computation of neural activity from fMRI brain imagery. Also, with this technique, shapes can be mapped to shperes for shape analysis, registration or other purposes. Our technique is based on conformal mappings which map genus-zero surface: in fmri case cortical or other surfaces, onto a sphere in an angle preserving manner. [[Projects:ConformalFlatteningRegistration|More...]]
| + | Our goal is to begin a longitudinal study of early brain development by cortical thickness in autistic children and controls (2 years with follow-up at 4 years). We want to be able to make statistical group comparisons. [[ProjectWeek200706:CorticalThicknessForAutism|More...]] |
| | | | |
| − | <font color="red">'''New: '''</font> Y. Gao, J. Melonakos, and A. Tannenbaum. Conformal Flattening ITK Filter. ISC/NA-MIC Workshop on Open Science at MICCAI 2006. | + | <font color="red">'''New: '''</font> Participation in the NA-MIC Summer 2007 Project Week. |
| | | | |
| | |} | | |} |
| − |
| |
| | == NA-MIC Kit == | | == NA-MIC Kit == |
| | | | |
| | === NAMIC Software Process === | | === NAMIC Software Process === |
| − | | + | # [[NA-MIC/Projects/NA-MIC_Kit/CMake_-_NAMIC_Kit_Building|CMake - NAMIC Kit Building]] (Kitware) |
| − | {| cellpadding="10"
| + | # [[NA-MIC/Projects/NA-MIC_Kit/CPack_-_NAMIC_Kit_Distribution|CPack - NAMIC Kit Distribution]] (Kitware) |
| − | | style="width:15%" |
| + | # [[NA-MIC/Projects/NA-MIC_Kit/Dart_2_and_CTest_-_Software_Quality|Dart 2 and CTest - Software Quality]] (GE, Kitware) |
| − | | style="width:85%" | | |
| − | | |
| − | |} | |
| | | | |
| | === Software Infrastructure === | | === Software Infrastructure === |
| − | | + | # [[NA-MIC/Projects/NA-MIC_Kit/Slicer3|Slicer 3]] (Isomics, GE, Kitware, UCSD, UCLA) |
| − | {| cellpadding="10"
| + | ## [[NA-MIC/Projects/NA-MIC_Kit/MRML|MRML]] (Isomics, Kitware, GE) |
| − | | style="width:15%" | | + | ## [[NA-MIC/Projects/NA-MIC_Kit/Coordinate_Systems|Coordinate Systems]] (GE, Isomics) |
| − | | style="width:85%" | | + | ## [[NA-MIC/Projects/NA-MIC_Kit/IO_Unification|IO Unification]] (GE, Isomics) |
| − | | + | ## [[NA-MIC/Projects/NA-MIC_Kit/Execution_Model|Execution Model]] (GE) |
| − | |} | + | ## [[NA-MIC/Projects/NA-MIC_Kit/Grid_Computing|Grid Computing]] (UCSD, GE, Kitware, Isomics) |
| | + | # [[NA-MIC/Projects/NA-MIC_Kit/Licenses_Unification|Licenses Unification]] (Kitware, Isomics) |
| | + | # Toolkits |
| | + | ## [[NA-MIC/Projects/NA-MIC_Kit/KWWidgets|KWWidgets]] (Kitware) |
| | + | ## [[NA-MIC/Projects/NA-MIC_Kit/Teem|Teem]] (Harvard/BWH) |
| | | | |
| | === Training & Dissemination === | | === Training & Dissemination === |
| − | | + | # [[NA-MIC/Projects/NA-MIC_Kit/Training_Material_and_Workshops_for_NA-MIC_Kit|Training Material and Workshops for NA-MIC Kit]] (MGH, BWH) |
| − | {| cellpadding="10"
| + | # [[NA-MIC/Projects/NA-MIC_Kit/Dissemination|Dissemination for NA-MIC]] (Isomics, Kitware, Harvard) |
| − | | style="width:15%" |
| |
| − | | style="width:85%" |
| |
| − | | |
| − | |} | |
| | | | |
| | == Other Projects == | | == Other Projects == |
| | | | |
| − | {| cellpadding="10"
| + | # [[Non_Rigid_Registration|Non-Rigid Registration]] |
| − | | style="width:15%" | [[Image:vxl.gif|200px]]
| + | # [[NA-MIC/Projects/Diffusion_Image_Analysis/FreeSurfer_NRRD_IO|FreeSurfer NRRD IO]] |
| − | | style="width:85%" |
| + | # [[Algorithm:MGH:FreeSurferNumericalRecipiesReplacement|FreeSurfer Numerical Recipies Replacement]] |
| − | | + | # [https://www.slicer.org/wiki/Slicer3:Fluorescence_and_Electron_Microscopy_Support Slicer3 Fluorescence and Electron Microscopy Support] |
| − | == [[Projects:FreeSurferNumericalRecipiesReplacement|Numerical Recipies Replacement]] ==
| |
| − | | |
| − | Our objective is to replace algorithms using the proprietary Numerical Recipes for C source base in FreeSurfer in the efforts to open-source FreeSurfer. This project has been completed through the use of the open source packages VXL (VNL) and Cephes. This includes the complete replacement of all Numerical Recipes in C code, and the implementation of a battery of unit tests for each replaced function. Currently the open source release is at a beta stage, and 25 beta releases of the source have been made. We anticipate a complete open source release in first quarter 2008. [[Projects:FreeSurferNumericalRecipiesReplacement|More...]]
| |
| − | | |
| − | <font color="red">'''New: '''</font> Completed
| |
| − | | |
| − | |}
| |