NAMIC Wiki:ShapeAnalysis:ShapeAnalysisIOstandardization
From NAMIC Wiki
Home < NAMIC Wiki:ShapeAnalysis:ShapeAnalysisIOstandardization
Contents
Unified format for Input Output of Studies/Population description for Shape Analysis data
General
- Currently there is no standard fileformat for describing shape analysis studies
- Is some of this data coming in Excel format? Do we need to be able to import that as well?
- At the simplest level, data is stored in comma/tab separated lists, rarely in excel. Part of the information on the staistical study description is somtimes supplied in excel (e.g. subject names, groups id's, age etc), the shape information so far though has never been stored/handled with Excel.
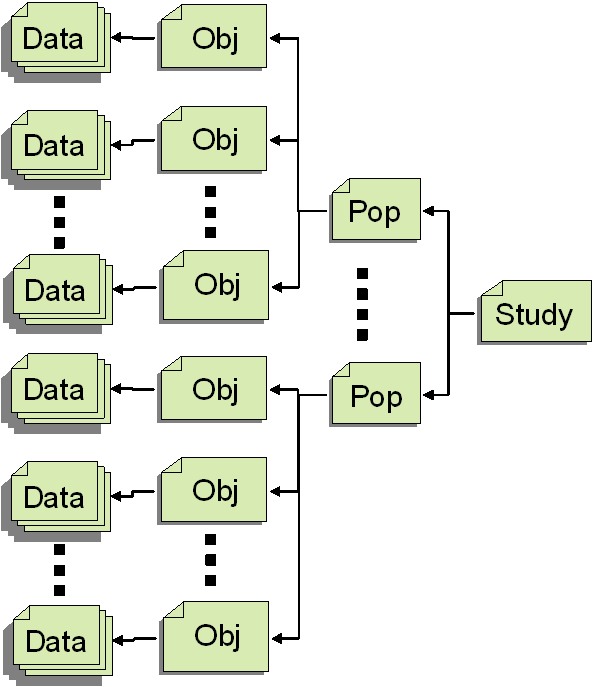
- Shape analysis studies are applied to a set of populations
- Each population is individually defined
- Subjects/objects can belong to multiple populations and studies
Base format for File IO
- As a base for storing the data, XML would serve best
- We need to balance the capabilities of XML against the simiplicity of a comma separated value format (CSV). For instance, if the objects or measurement could change from experiment to experiment, then an XML format is a good choice. I would suggest having a generic "measurement" tag that has an attribute specifying the measurement "name". This provides a great deal of flexibility.
- XML allows ASCII readability/editability
- XML-editors allow easy editing
- Future changes to the fileformat are easier to handle using an xml based fileformat
- Indeed the objects can change from experiment to experiment. E.g. in Namic we have the Boston VA data. For the same subjects, there are segmentations of several structures. We want to analyze each one of them without the need to setup a separate study file for each analysis. But rather one study file should be enough, and then simply different objects are selected from the subject info. But it goes even further, in that there could per object multiple measurement/shape desccriptions, depending on choice of scale (often we statistically analyze an anatomical structure in its original scale, in unit scale and in brain-size normalized scale).
- The current version has objects with freely choosable names, and each object has any number of attributes/measurements. The name of the measurement has to be a keyword that needs to be given to a statistical processing filter.
- In regard to the naming of the attributes/measurement, we should agree on a large set of predefined measurements, such that people call the same thing the same way, but additionally allow for anybody to create their own measurement keywords. I don't think though that we need a special measurement tag, as a measurement is worth little to somebody who does not know its meaning.
- Care must be taken in designing the XML format. Terseness and generality are good goals.
- Which XML reader? Use ITK XML reader, libxml2?
- ITK already has expat included. This is a light weight XML parser.
- The current implementation uses libxml2, but this should be easy to change (but not really a priority for me right now), as the classes were designed for easy pluggable xml libraries, i.e. there is an additional abstract xmlIO class between Study/Population/Subject representation and the xml implementation. One onlys need to write a specific implementation of the xmlIO base class.
- All description should be easily extendable, i.e. all keys-value pairs are read in, and user determines which pairs are needed.
- All information should be stored in a single central file
- The loaded classes should be organized in a hierarchical fashion
Base format for Internal representation
- Classes compatible to itk representations: DataObject based
- Predefined access of the main data through direct functions
- Support of query functions for access of data
- For example: Query(NC,object1) returns a list of all subject id's that in the NC population with a object1 object
- All referenced data (images, surfaces) are not loaded when loading the population file, but have to be loaded by the user
- Reader Writer is done through ImageIO, and thus can be easily extended to CVS and other formats
Information stored on the population & subject level, models, features
- specs for Study, Population and Subject data
- NAMIC_Wiki:ShapeAnalysis:StatModel_format
- specs for Feature & Statistics data
Additional Material
- Older Proposal for XML File hierarchy shown on a hippocampus shape analysis case:File:02-05-NAMIC-ShapeAnalysisStandard.ppt, use only for nostalgic reasons
- New Proposal/Example for XML File hierarchy: File:TestStudy.xml.gz
- No abbreviations in keywords
- Designation of type (only default support for standard types, but extensible)
- No numbering of populations and subjects (identified through ID)
- Base XML tag StatisticalStudyDescription
- Patient parameters have 2 entries, one for the value and a descriptive entry
- Schematic DOM model