NCI Evaluating NA-MIC Tools for Image Analysis
From NAMIC Wiki
Home < NCI Evaluating NA-MIC Tools for Image Analysis
 Return to Project Week Main Page |
Key Investigators
- NCI: Yanling Liu, Christopher Kurcz
- KnowledgeVis: Curt Lisle
Objective
The National Cancer Institute performs research in multiple internal laboratories. Microscopy, small animal imaging, and other cancer-fighting work may all benefit from increased use of imaging technology. The purpose of this project is to apply NA-MIC tools and technology to research datasets.
Approach, Plan
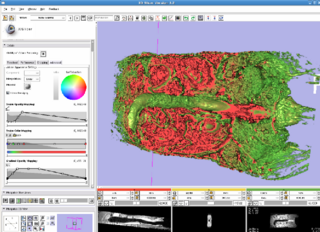
- We will bring sample datasets from microscopy and small animal scans to the project week. These datasets are in ICS (microscopy format similar to NRRD), and MRI datasets of mice.
- During the week, we will use Slicer to adjust, register, segment, and analyze these datasets. Our goal is to determine which modules are the most useful for biological analysis, and contribute to the testing and refinement of those modules.
Progress
- We consulted with Stephen Alyward about the datasets and learned to perform registration on labelmaps. We will follow up with Luis to consider work with the explicit snakes algorithm for these datasets.
- We used Slicer to read anatomical datasets of vasculature near turmors for vessel 3D modeling and analysis for angiogenesis studies.
- There is still header data missing in the DICOM created from data imports, but this can be solved by carrying the tag information through the MRML nodes using key/value pairs.
- Development on a new module for cell wall segmentation was started.