Difference between revisions of "OpenIGTLink OldPage"
(Created page with ' OpenIGTLink provides a standardized mechanism for communications among computers and devices in operating rooms (OR) for wide variety of image-guided therapy (IGT) applications…') |
m (Text replacement - "http://www.slicer.org/slicerWiki/index.php/" to "https://www.slicer.org/wiki/") |
||
| (One intermediate revision by one other user not shown) | |||
| Line 36: | Line 36: | ||
==Software == | ==Software == | ||
| − | * [ | + | * [https://www.slicer.org/wiki/Modules:OpenIGTLinkIF Interface for 3D Slicer] |
* [[OpenIGTLink/Simlators | Simulator programs]] | * [[OpenIGTLink/Simlators | Simulator programs]] | ||
| Line 46: | Line 46: | ||
**Tutorials on image-guided therapy (IGT) using Slicer 3 and OpenIGTLink are available at [[IGT:ToolKit]] | **Tutorials on image-guided therapy (IGT) using Slicer 3 and OpenIGTLink are available at [[IGT:ToolKit]] | ||
** [[OpenIGTLink/IGSTK | Interface for Image-Guided Surgery Toolkit (IGSTK)]] | ** [[OpenIGTLink/IGSTK | Interface for Image-Guided Surgery Toolkit (IGSTK)]] | ||
| − | **[ | + | **[https://www.slicer.org/wiki/Modules:OpenIGTLinkIF-Documentation-3.6 Tutorial on 3D Slicer and Tracker simulator in 3D Slicer OpenIGTLink IF Documentation page.] |
** [[OpenIGTLink/ProtocolV2/Trial| How to use the new features in Version 2 in 3D Slicer.]] | ** [[OpenIGTLink/ProtocolV2/Trial| How to use the new features in Version 2 in 3D Slicer.]] | ||
| Line 77: | Line 77: | ||
*Related web pages | *Related web pages | ||
| − | ** [ | + | ** [https://www.slicer.org/wiki/Slicer3:_Image_Guided_Therapy_(IGT) Slicer 3 Image-Guided Therapy (IGT) suite] |
| − | ** [ | + | ** [https://www.slicer.org/wiki/Slicer3:Slicer_Daemon Slicer 3 Daemon] Another network interface in 3D Slicer |
** [http://www.opendmtp.org/ OpenDMTP] - Open Device Monitoring and Tracking Protocol | ** [http://www.opendmtp.org/ OpenDMTP] - Open Device Monitoring and Tracking Protocol | ||
** [http://en.wikipedia.org/wiki/Uniform_Driver_Interface Uniform Driver Interface (UDI)] | ** [http://en.wikipedia.org/wiki/Uniform_Driver_Interface Uniform Driver Interface (UDI)] | ||
| Line 87: | Line 87: | ||
<hr/> | <hr/> | ||
== License== | == License== | ||
| − | [http:// | + | [http://opensource.org/licenses/BSD-3-Clause New BSD license] ©2008 Insight Software Consortium<br /> |
== Mailing List / Contact == | == Mailing List / Contact == | ||
Latest revision as of 17:15, 10 July 2017
Home < OpenIGTLink OldPage
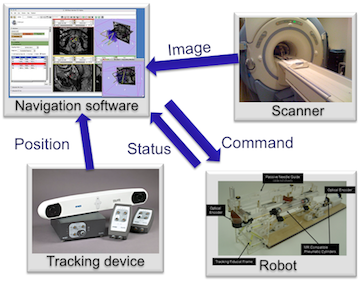
OpenIGTLink provides a standardized mechanism for communications among computers and devices in operating rooms (OR) for wide variety of image-guided therapy (IGT) applications.
Examples of such applications include:
- Stereotactic surgical guidance using optical position sensor and medical image visualization software.
- Intraoperative image guidance using real-time MRI and medical image visualization software
- Robot-assisted intervention using robotic device and surgical planning software
See Applications section below for a list of research projects using OpenIGTLink.
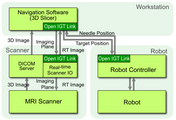
OpenIGTLink is a set of digital messaging formats and rules (protocol) used for data exchange on a local area network (LAN). The specification of OpenIGTLink and its reference implementation, the OpenIGTLink Library, are available free of charge for any purpose including commercial use. An OpenIGTLink interface is available in popular medical image processing and visualization software 3D Slicer.
Protocol
OpenIGTLink Version 2
Extended Protocols
Downloads
Reference Library
Software
Tutorials/Events
- Tutorial Materials
- Tutorials on image-guided therapy (IGT) using Slicer 3 and OpenIGTLink are available at IGT:ToolKit
- Interface for Image-Guided Surgery Toolkit (IGSTK)
- Tutorial on 3D Slicer and Tracker simulator in 3D Slicer OpenIGTLink IF Documentation page.
- How to use the new features in Version 2 in 3D Slicer.
- Meetings / Workshops
Applications
OpenIGTLink has been used in a number of research projects. Here are some examples:
Please see the list of hardware that supports the OpenIGTLink Protocol.
References
- Studies using OpenIGTLink
- Tokuda J, Fischer GS, Papademetris X, Yaniv Z, Ibáñez L, Cheng P, Liu H, Blevins J, Arata J, Golby AJ, Kapur T, Pieper S, Burdette EC, Fichtinger G, Tempany CM, Hata N. OpenIGTLink: an open network protocol for image-guided therapy environment. Int J Med Robot. 2009 Dec;5(4):423-34.
- Arata J, Kozuka H, Kim HW, Takesue N, Vladimirov B, Sakaguchi M, Tokuda J, Hata N, Chinzei K, Fujimoto H. Open core control software for surgical robots. Int J Comput Assist Radiol Surg. 2010 May;5(3):211-20. Epub 2009 Jul 28.
- Tokuda J, Fischer GS, DiMaio SP, Gobbi DG, Csoma C, Mewes PW, Fichtinger G, Tempany CM, Hata N. Integrated navigation and control software system for MRI-guided robotic prostate interventions. Comput Med Imaging Graph. 2010 Jan;34(1):3-8. Epub 2009 Aug 20.
- Elhawary H, Liu H, Patel P, Norton I, Rigolo L, Papademetris X, Hata N, Golby AJ. Intraoperative real-time querying of white matter tracts during frameless stereotactic neuronavigation. Neurosurgery. 2011 Feb;68(2):506-16.
- Egger J, Tokuda J, Chauvin L, Freisleben B, Nimsky C, Kapur T, Wells W. Integration of the OpenIGTLink Network Protocol for image-guided therapy with the medical platform MeVisLab, Int J Med Robot Article first published online: 28 FEB 2012 DOI: 10.1002/rcs.1415
- Related web pages
- Slicer 3 Image-Guided Therapy (IGT) suite
- Slicer 3 Daemon Another network interface in 3D Slicer
- OpenDMTP - Open Device Monitoring and Tracking Protocol
- Uniform Driver Interface (UDI)
- Debian Source Package: OpenIGTLink
Support
License
New BSD license ©2008 Insight Software Consortium
Mailing List / Contact
We are launching the OpenIGTLink mailing list (openigtlink@bwh.harvard.edu). If you would like to join, please subscribe from OpenIGTLink ML Info Page or send Junichi Tokuda (tokuda at bwh.harvard.edu) a message with:
- Your name
- e-mail address you would like to use to post and receive ML articles on
- Affiliation
- Your research topic
Acknowledgements
- Contributors
- Luis Ibáñez, Ph.D., Kitware, Inc.
- Junichi Tokuda, Ph.D., BWH
- Csaba Csoma, John's Hopkins University
- Jack Blevins, Acoustic MedSystems, Inc.
- Patrick Cheng, Ph.D., Georgetown University
- Haying Liu, Brigham and Women's Hospital
- Kiyoyuki Chinzei, Ph.D., National Institute of Advanced Industrial Science and Technology, Japan
- Jumpei Arata, Ph.D., Nagoya Institute of Technology, Japan
- Steve Pieper, Ph.D., Isomics, Inc.
- Yuichiro Hayashi, Ph.D., Nagoya University, Japan
- Atsushi Yamada, Ph.D., Nagoya Institute of Technology, Japan
- Insight Software Consortium
- Funding Support
- Enabling Technologies for MRI-Guided Prostate Interventions funded by NIH (PI: Clare Tempany, MD)
- National Center for Image-Guided Therapy (NCIGT)
- National Alliance for Medical Image Computing (NA-MIC)
- Intelligent Surgical Instrument Project sponsored by The Ministry of International Trade and Industry, Japan