Difference between revisions of "Problems and Solutions on SPL Machine Blog"
| Line 168: | Line 168: | ||
And everything runs fine... | And everything runs fine... | ||
| + | |||
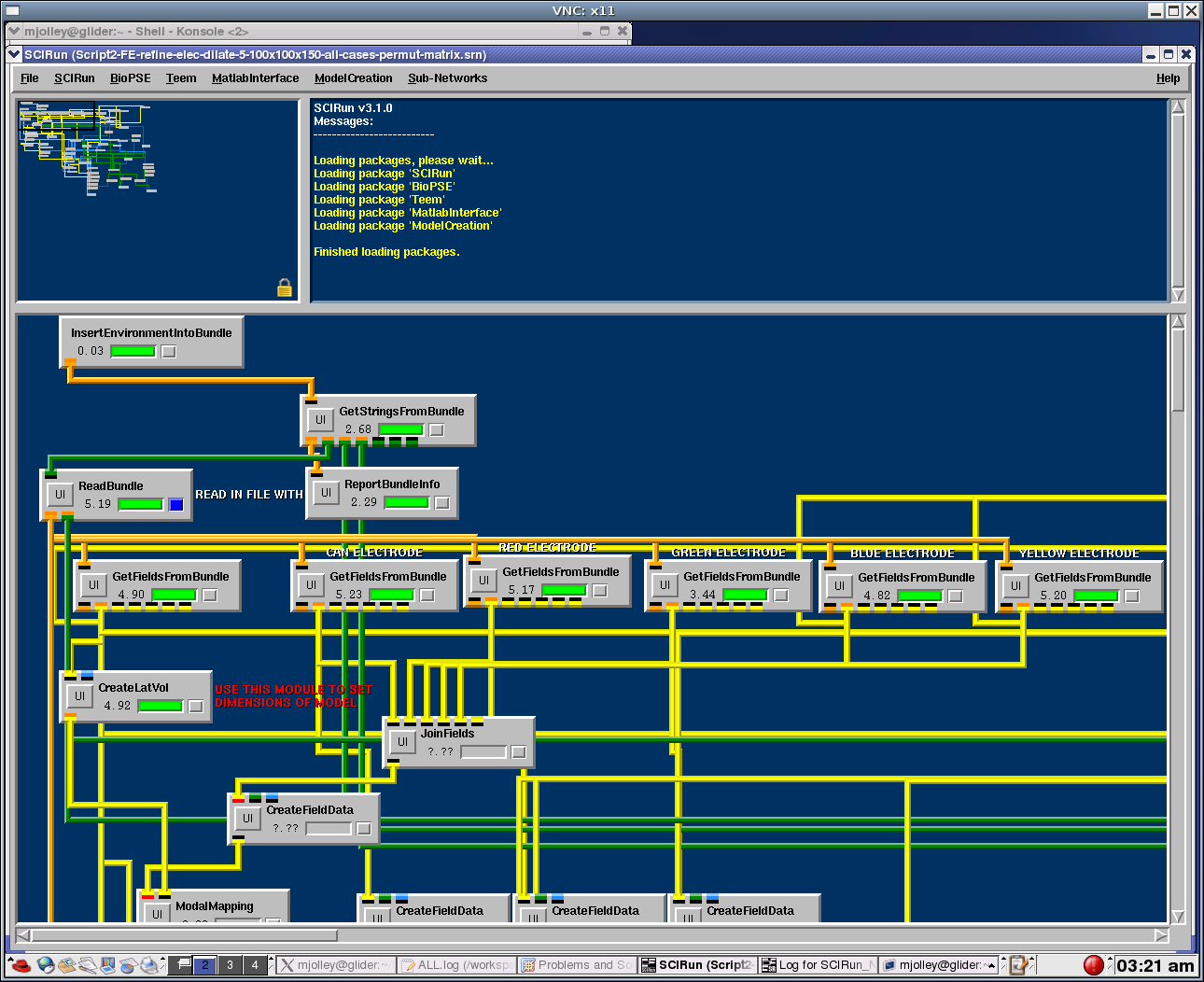
| + | I checked and all the bundles in GetFieldsFromBundle are Defined, it just won't run the JoinField. Is there some environmental variable that is not set and does not allow this to run? | ||
| + | [[Image:VNC2]] | ||
--[[User:Mjolley|Mjolley]] 11:51, 21 August 2007 (EDT) | --[[User:Mjolley|Mjolley]] 11:51, 21 August 2007 (EDT) | ||
Revision as of 19:12, 21 August 2007
Home < Problems and Solutions on SPL Machine BlogCurrent Problems on Debugging for SCIRun on SPL Machines
With the install of Fedora 7 we have been having some problems, namely with the OpenGL driver recognition by SCIRun.
For example when we utilize the tools on the previous page SCIRun gets pointed at the correct drivers as shown here:
spl_tm64_1:/workspace/mjolley/Modeling/trunk/SCIRun/bin% ldd scirun | grep GL
libGL.so.1 => /usr/lib64/nvidia/libGL.so.1 (0x0000003f8e200000)
libGLU.so.1 => /usr/lib64/libGLU.so.1 (0x000000360c200000)
libGLcore.so.1 => /usr/lib64/nvidia/libGLcore.so.1 (0x0000003f80e00000)
When these drivers are recognized SCIRun runs as expected. However, each time SCIRun is run it reverts back to the wrong drivers as evidenced here.
spl_tm64_1:/workspace/mjolley/Modeling/trunk/SCIRun/bin% ldd scirun | grep GL
libGL.so.1 => /usr/lib64/libGL.so.1 (0x0000003f84800000)
libGLU.so.1 => /usr/lib64/libGLU.so.1 (0x000000360c200000)
After this, any OpenGL dependent modules crash upon opening. If you repeat the steps "unsetenv" and run Dav's script again you get back to:
spl_tm64_1:/workspace/mjolley/Modeling/trunk/SCIRun/bin% ldd scirun | grep GL
libGL.so.1 => /usr/lib64/nvidia/libGL.so.1 (0x0000003f8e200000)
libGLU.so.1 => /usr/lib64/libGLU.so.1 (0x000000360c200000)
libGLcore.so.1 => /usr/lib64/nvidia/libGLcore.so.1 (0x0000003f80e00000)
To summarize:
unset LD_LIBRARY_PATH
ldd scirun | grep GL
<correct GL is reported>
run script
ldd scirun | grep GL
<wrong GL is reported>
So I put:
unset LD_LIBRARY_PATH into the first line of the script(run in bash) and into my .bashrc and I still have the same behavior where it switches back from the good OpenGL setup to the mesa drivers after initially being pointed to the correct ones.
This still didn't seem to do it so we put in the following line in the script:
create_scirun_script() {
echo "scirun -E ${NETWORK} --logfile ALL.log" >/tmp/script-fe.sh
chmod 0770 /tmp/script-fe.sh
}
to
create_scirun_script() {
echo "unset LD_LIBRARY_PATH" > /tmp/script-fe.sh
echo "scirun -E ${NETWORK} --logfile ALL.log" >> /tmp/script-fe.sh
chmod 0770 /tmp/script-fe.sh
}
And the above behavior with it switching back to the MESA drivers appears to have stopped, but the script still always hangs at JoinField within the first three bundles run in the net by the script. I went back and confirmed this set of .bdl files run without errors on my ubuntu machine and in the manual nets on the SPL machine. So it must be an environmental variable. It typically hangs in the first, second, or third run in Scripts on the SPL machines always at the same position. I am not sure what is up as there is not a good error message:
This is the output in the command line script window:
spl_tm64_1:/workspace/mjolley% bash bash-3.2$ export PATH=$PATH:/workspace/mjolley/Modeling/trunk/SCIRun/bin bash-3.2$ ./SCIRun_Scripts/run_all.sh /projects/cardio/Clinical-HClean/ /projects/cardio/Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn Results/Clinical ls: cannot access /projects/cardio/Clinical-HClean//Electrodes_Plus_Torso/2ybdls//Four/*.bdl: No such file or directory cat: /tmp/idx1: No such file or directory cat: /tmp/idx2: No such file or directory cat: /tmp/idx3: No such file or directory cat: /tmp/idx4: No such file or directory ls: cannot access /projects/cardio/Clinical-HClean//Electrodes_Plus_Torso/10ybdls//Four/*.bdl: No such file or directory ldd: ./scirun: No such file or directory Parsed .scirunrc... /home/mjolley/.scirunrc Loading Tcl,Tk,tk, Itcl,Itk,Blt,Widgets loading scirun network file: /projects/cardio/Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn scirun> loading file: SCIRun_Scripts/Permutations/P1-500-0.mat loading file: /projects/cardio/Clinical-HClean//Electrodes_Plus_Torso/10ybdls//One/10y-Left-abd-can+10cm-right-parasternal-T4-top.bdl Compiling: ArrayObjectFieldCreateAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double >,vector<double > > > Compiling: ArrayObjectFieldCreateAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double >,vector<double > > > Compiling: ArrayObjectFieldCreateAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double >,vector<double > > > Compiling: ArrayObjectFieldCreateAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double >,vector<double > > > Compiling: ArrayObjectFieldCreateAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double >,vector<double > > > Compiling: ArrayObjectFieldDataScalarAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > ,TetVolMesh<TetLinearLgn<Point> > ::Cell> Compiling: ArrayObjectFieldDataScalarAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > ,TetVolMesh<TetLinearLgn<Point> > ::Cell> Compiling: Compiling: ArrayObjectFieldDataScalarAlgoTMergeFieldsAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > ,TetVolMesh<TetLinearLgn<Point> > ::Cell<> GenericField<TetVolMesh<TetLinearLgn<Point> > ,NoDataBasis<double> ,vector<double> > > Compiling: ArrayObjectFieldLocationElemAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > > Compiling: ArrayObjectFieldDataScalarAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > ,TetVolMesh<TetLinearLgn<Point> > ::Cell> Compiling: ArrayObjectFieldLocationElemAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > > Compiling: ArrayObjectFieldDataScalarAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > ,TetVolMesh<TetLinearLgn<Point> > ::Cell> Compiling: ArrayObjectFieldLocationElemAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > > Compiling: ArrayObjectFieldLocationElemAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > > Compiling: ArrayObjectFieldLocationElemAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > > Compiling: ArrayObjectFieldElemVolumeAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > ,TetVolMesh<TetLinearLgn<Point> > ::Cell> Compiling: ArrayObjectFieldElemVolumeAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > ,TetVolMesh<TetLinearLgn<Point> > ::Cell> Compiling: ArrayObjectFieldElemVolumeAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > ,TetVolMesh<TetLinearLgn<Point> > ::Cell> Compiling: ArrayObjectFieldElemVolumeAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > ,TetVolMesh<TetLinearLgn<Point> > ::Cell> Compiling: ArrayObjectFieldElemVolumeAlgoT<GenericField<TetVolMesh<TetLinearLgn<Point> > ,ConstantBasis<double> ,vector<double> > ,TetVolMesh<TetLinearLgn<Point> > ::Cell> Compiling: ALGOArrayEngine_1586526061_LCELID_FS<double> Compiling: ALGOArrayEngine_1586525968_LCELID_FS<double> Compiling: ALGOArrayEngine_1586526092_LCELID_FS<double> Compiling: ALGOArrayEngine_1586525999_LCELID_FS<double>
Then SCIRun hangs...
In comparison on my ubuntu machine it is not nearly as verbose regarding compilation etc running the same script...
mj@mj-quad-desktop:~$ SCIRun_Scripts/run_all.sh Clinical-HClean/ Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn Results/Clinical ls: Clinical-HClean//Electrodes_Plus_Torso/2ybdls//Four/*.bdl: No such file or directory cat: /tmp/idx1: No such file or directory cat: /tmp/idx2: No such file or directory cat: /tmp/idx3: No such file or directory cat: /tmp/idx4: No such file or directory ls: Clinical-HClean//Electrodes_Plus_Torso/10ybdls//Four/*.bdl: No such file or directory Parsed .scirunrc... .scirunrc Loading Tcl,Tk,tk, Itcl,Itk,Blt,Widgets loading scirun network file: Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn NETFILE = Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn NETFILE = /home/mj/Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn filename=Clinical-HClean//Electrodes_Plus_Torso/10ybdls//One/10y-Left-abd-can+25cm-sc-left-T10back.bdl filename=Clinical-HClean//Electrodes_Plus_Torso/10ybdls//One/10y-Left-abd-can+25cm-sc-left-T10back.bdl filename=SCIRun_Scripts/Permutations/P1-500-0.mat filename=SCIRun_Scripts/Permutations/P1-500-0.mat scirun> loading file: SCIRun_Scripts/Permutations/P1-500-0.mat loading file: Clinical-HClean//Electrodes_Plus_Torso/10ybdls//One/10y-Left-abd-can+10cm-right-parasternal-T4-top.bdl Creating /home/mj/Results/Clinical/mj-21-Aug/ Creating /home/mj/Results/Clinical/mj-21-Aug/ Creating /home/mj/Results/Clinical/mj-21-Aug/ saving file: /home/mj/Results/Clinical/mj-21-Aug/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix-10y-Left-abd-can+10cm-right-parasternal-T4-top-P1-500-0.txt Parsed .scirunrc... .scirunrc Loading Tcl,Tk,tk, Itcl,Itk,Blt,Widgets loading scirun network file: Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn NETFILE = Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn NETFILE = /home/mj/Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn filename=Clinical-HClean//Electrodes_Plus_Torso/10ybdls//One/10y-Left-abd-can+25cm-sc-left-T10back.bdl filename=Clinical-HClean//Electrodes_Plus_Torso/10ybdls//One/10y-Left-abd-can+25cm-sc-left-T10back.bdl filename=SCIRun_Scripts/Permutations/P1-500-0.mat filename=SCIRun_Scripts/Permutations/P1-500-0.mat scirun> loading file: Clinical-HClean//Electrodes_Plus_Torso/10ybdls//One/10y-Left-abd-can+10cm-right-parasternal-T6-top.bdl loading file: SCIRun_Scripts/Permutations/P1-500-0.mat saving file: /home/mj/Results/Clinical/mj-21-Aug/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix-10y-Left-abd-can+10cm-right-parasternal-T6-top-P1-500-0.txt saving file: /home/mj/Results/Clinical/mj-21-Aug/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix-10y-Left-abd-can+10cm-right-parasternal-T6-top-P1-500-0.db.txt saving file: /home/mj/Results/Clinical/mj-21-Aug/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix-10y-Left-abd-can+10cm-right-parasternal-T6-top-P1-500-0.bdl Parsed .scirunrc... .scirunrc Loading Tcl,Tk,tk, Itcl,Itk,Blt,Widgets loading scirun network file: Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn NETFILE = Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn NETFILE = /home/mj/Clinical-HClean/SCIRun_Nets/Script2_FEM/Script2-FE-refine-elec-dilate-5-100x100x150-all-cases-permut-matrix.srn filename=Clinical-HClean//Electrodes_Plus_Torso/10ybdls//One/10y-Left-abd-can+25cm-sc-left-T10back.bdl filename=Clinical-HClean//Electrodes_Plus_Torso/10ybdls//One/10y-Left-abd-can+25cm-sc-left-T10back.bdl filename=SCIRun_Scripts/Permutations/P1-500-0.mat filename=SCIRun_Scripts/Permutations/P1-500-0.mat scirun> loading file: SCIRun_Scripts/Permutations/P1-500-0.mat loading file: Clinical-HClean//Electrodes_Plus_Torso/10ybdls//One/10y-Left-abd-can+25cm-sc-left-T10back.bdl
And everything runs fine...
I checked and all the bundles in GetFieldsFromBundle are Defined, it just won't run the JoinField. Is there some environmental variable that is not set and does not allow this to run? File:VNC2
--Mjolley 11:51, 21 August 2007 (EDT)