Difference between revisions of "ProjectWeek200706:vtkITKWrapperForRuleBasedSegmentation"
| Line 1: | Line 1: | ||
{| | {| | ||
|[[Image:ProjectWeek-2007.png|thumb|320px|Return to [[2007_Programming/Project_Week_MIT|Project Week Main Page]] ]] | |[[Image:ProjectWeek-2007.png|thumb|320px|Return to [[2007_Programming/Project_Week_MIT|Project Week Main Page]] ]] | ||
| − | |[[Image: | + | |[[Image:Dlpfc-slicer.png|thumb|120px|Scatter plot of the original FA data through the genu of the corpus callosum of a normal brain.]] |
| − | |||
|} | |} | ||
__NOTOC__ | __NOTOC__ | ||
===Key Investigators=== | ===Key Investigators=== | ||
| − | *Georgia Tech: John Melonakos | + | * Georgia Tech: John Melonakos, Ramsey Al-Hakim |
* Kitware: Brad Davis | * Kitware: Brad Davis | ||
* BWH: Marek Kubicki | * BWH: Marek Kubicki | ||
| + | * UCI: Jim Fallon | ||
<div style="margin: 20px;"> | <div style="margin: 20px;"> | ||
| Line 15: | Line 15: | ||
<div style="width: 27%; float: left; padding-right: 3%;"> | <div style="width: 27%; float: left; padding-right: 3%;"> | ||
<h1>Objective</h1> | <h1>Objective</h1> | ||
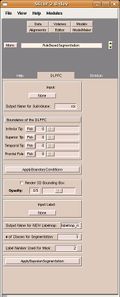
| − | We are developing | + | We are developing rule-based segmentation techniques which speedup the process and improve the accuracy for delineating the DLPFC in brain MRI scans. Our objective is to develop Slicer modules to facilitate clinical use of these techniques. |
| − | |||
| + | A functional Slicer2 module has been developed and now needs to be tested and used by our Core 3 partners. Furthermore, in order to provide continued support for this module, we will port this code to Slicer3. | ||
</div> | </div> | ||
| Line 23: | Line 23: | ||
<h1>Approaches and Challenges </h1> | <h1>Approaches and Challenges </h1> | ||
| − | + | Our approach for segmenting the DLPFC is described in the references below. The challenge is to make this software user-friendly to enable clinical use of the tool. | |
| − | Our approach for | ||
</div> | </div> | ||
| Line 35: | Line 34: | ||
This is where you put in progress made in Project Week 2007. | This is where you put in progress made in Project Week 2007. | ||
| − | ==== | + | ====2005-2007==== |
| − | + | This code was developed between 2005-2007. First is was developed and tested in Matlab. Then the sub-volume creation rules were ported to Slicer2 while the Bayesian segmentation was ported to ITK (see the references below for more detail). Finally, in early 2007, a vtk wrapper of the ITK Bayesian code was developed, thus completing the Slicer2 RuleBasedSegmentation module. | |
</div> | </div> | ||
| Line 46: | Line 45: | ||
===Publications=== | ===Publications=== | ||
| − | * | + | * Ramsey Al-Hakim, James Fallon, Delphine Nain, John Melonakos, and Allen Tannenbaum. A dorsolateral prefrontal cortex semi-automatic segmenter. In SPIE Medical Imaging, 2006. |
| − | * | + | * J. Melonakos, K. Krishnan, and A. Tannenbaum. An ITK Filter for Bayesian Segmentation: itkBayesianClassifierImageFilter. Insight Journal, 2006. |
| − | * | + | * J. Melonakos, R. Al-Hakim, J. Fallon, and A. Tannenbaum. Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit. Insight Journal, 2005. |
| − | |||
Revision as of 02:07, 25 May 2007
Home < ProjectWeek200706:vtkITKWrapperForRuleBasedSegmentation Return to Project Week Main Page |
Key Investigators
- Georgia Tech: John Melonakos, Ramsey Al-Hakim
- Kitware: Brad Davis
- BWH: Marek Kubicki
- UCI: Jim Fallon
Objective
We are developing rule-based segmentation techniques which speedup the process and improve the accuracy for delineating the DLPFC in brain MRI scans. Our objective is to develop Slicer modules to facilitate clinical use of these techniques.
A functional Slicer2 module has been developed and now needs to be tested and used by our Core 3 partners. Furthermore, in order to provide continued support for this module, we will port this code to Slicer3.
Approaches and Challenges
Our approach for segmenting the DLPFC is described in the references below. The challenge is to make this software user-friendly to enable clinical use of the tool.
Progress
June 2007 Project Week
This is where you put in progress made in Project Week 2007.
2005-2007
This code was developed between 2005-2007. First is was developed and tested in Matlab. Then the sub-volume creation rules were ported to Slicer2 while the Bayesian segmentation was ported to ITK (see the references below for more detail). Finally, in early 2007, a vtk wrapper of the ITK Bayesian code was developed, thus completing the Slicer2 RuleBasedSegmentation module.
Publications
- Ramsey Al-Hakim, James Fallon, Delphine Nain, John Melonakos, and Allen Tannenbaum. A dorsolateral prefrontal cortex semi-automatic segmenter. In SPIE Medical Imaging, 2006.
- J. Melonakos, K. Krishnan, and A. Tannenbaum. An ITK Filter for Bayesian Segmentation: itkBayesianClassifierImageFilter. Insight Journal, 2006.
- J. Melonakos, R. Al-Hakim, J. Fallon, and A. Tannenbaum. Knowledge-Based Segmentation of Brain MRI Scans Using the Insight Toolkit. Insight Journal, 2005.