Difference between revisions of "Project Week 25/Wrist Kinematics:"
Jfishbaugh (talk | contribs) |
|||
| (6 intermediate revisions by the same user not shown) | |||
| Line 19: | Line 19: | ||
|<!-- Approach and Plan bullet points --> | |<!-- Approach and Plan bullet points --> | ||
| + | |||
| + | * We will use Slicer and various tools for visualization, segmentation, and analysis of the dynamic MR sequence. | ||
|<!-- Progress and Next steps (fill out at the end of project week), bullet points --> | |<!-- Progress and Next steps (fill out at the end of project week), bullet points --> | ||
| − | ** 3D segmentation of individual bones was solved via level-set segmentation within itkSNAP. Preliminary tests with Slicer GrowCut were not satisfactory due to highly non-isotropic voxels, but more tests will be necessary to explore new advanced features within "Segment Editor". | + | * Slicer was used to import the DICOM sequence and export 10 individual volumes. The sequences module combined with volume rendering within Slicer provided exploratory qualatative analysis of the wrist kinematics. |
| + | |||
| + | * The 10 images in the sequence were used to estimate a template image with [http://stnava.github.io/ANTs/ ANTs]. | ||
| + | |||
| + | * 3D segmentation of individual bones was solved via level-set segmentation within [http://www.itksnap.org/pmwiki/pmwiki.php itkSNAP]. Preliminary tests with Slicer GrowCut were not satisfactory due to highly non-isotropic voxels, but more tests will be necessary to explore new advanced features within "Segment Editor". | ||
| + | |||
| + | * Spatiotemporal modeling was performed jointly with all bones using [https://github.com/jamesfishbaugh/shape4D shape4d]. | ||
| + | |||
|} | |} | ||
==Illustrations== | ==Illustrations== | ||
| + | |||
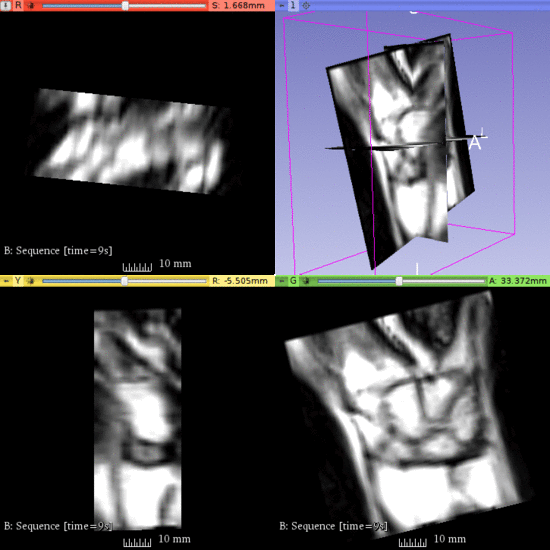
| + | Left) One observation of the wrist. Right) A sequence of volume renderings within Slicer to show the 10 observations. | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 32: | Line 43: | ||
<br> | <br> | ||
| + | |||
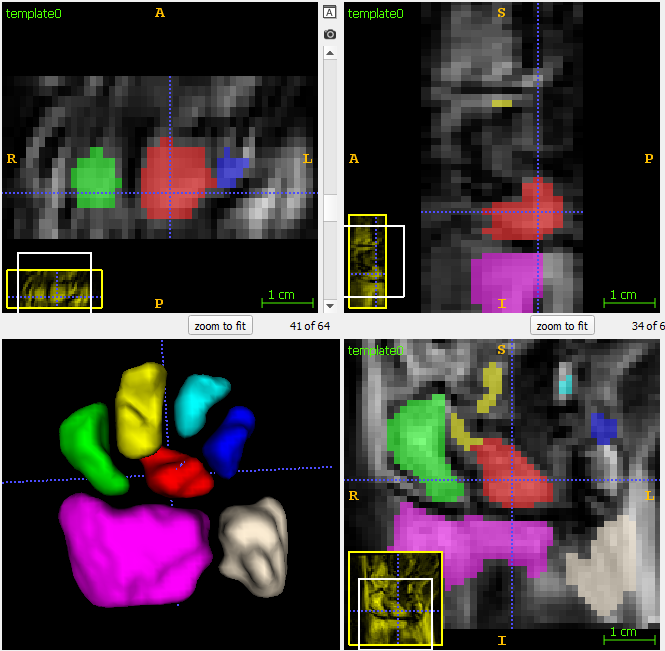
| + | Segmentation of the wrist bones in the template space, which are propagated back to the individual observations via mappings computed during template estimation. | ||
[[File:Wrist_template_seg.png]] | [[File:Wrist_template_seg.png]] | ||
<br> | <br> | ||
| + | |||
| + | Continous change trajectories via spatiotemporal modeling. Color denotes speed and vectors denote direction of change. | ||
http://research.engineering.nyu.edu/~fishbaugh/docs/wrist_quick.gif http://research.engineering.nyu.edu/~fishbaugh/docs/wrist_velocity_vectors.gif | http://research.engineering.nyu.edu/~fishbaugh/docs/wrist_quick.gif http://research.engineering.nyu.edu/~fishbaugh/docs/wrist_velocity_vectors.gif | ||
==Background and References== | ==Background and References== | ||
| + | [1] Fishbaugh, J., Durrleman, S., Prastawa, M., Gerig, G. Geodesic shape regression with multiple geometries and sparse parameters. Medical Image Analysis. Vol 39. pp. 1-17. (2017) | ||
| + | |||
| + | [2] Fishbaugh, J., Durrleman, S., Gerig, G. Estimation of smooth growth trajectories with controlled acceleration from time series shape data. Proc. of Medical Image Computing and Computer Assisted Intervention (MICCAI '11). Vol 6892, pp. 401-408. (2011) | ||
| + | |||
<!-- Use this space for information that may help people better understand your project, like links to papers, source code, or data --> | <!-- Use this space for information that may help people better understand your project, like links to papers, source code, or data --> | ||
Latest revision as of 10:08, 30 June 2017
Home < Project Week 25 < Wrist Kinematics:Back to Projects List
Key Investigators
- James Fishbaugh (NYU Tandon School of Engineering, USA)
- Guido Gerig (NYU Tandon School of Engineering, USA)
Project Description
| Objective | Approach and Plan | Progress and Next Steps |
|---|---|---|
|
|
|
Illustrations
Left) One observation of the wrist. Right) A sequence of volume renderings within Slicer to show the 10 observations.
 |
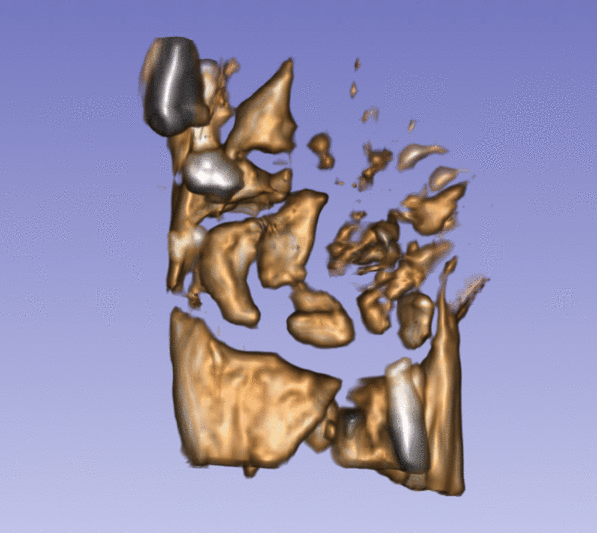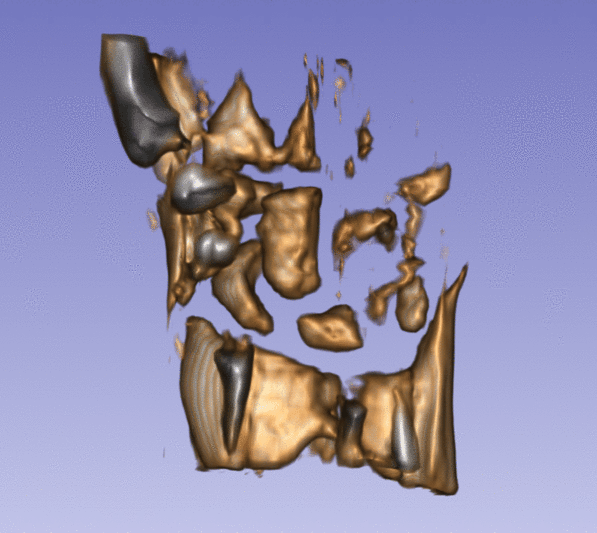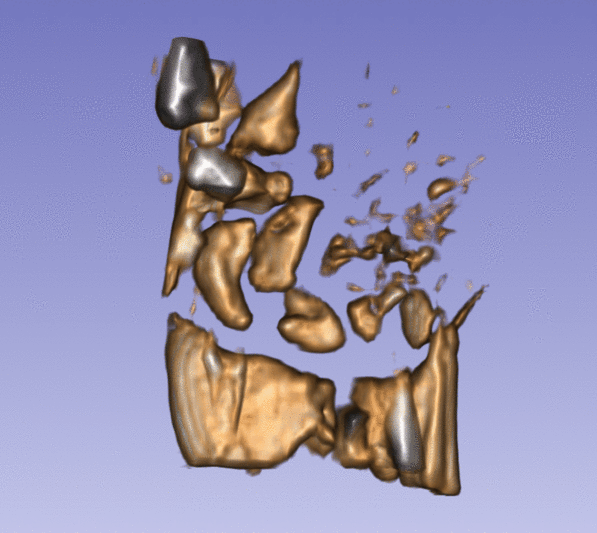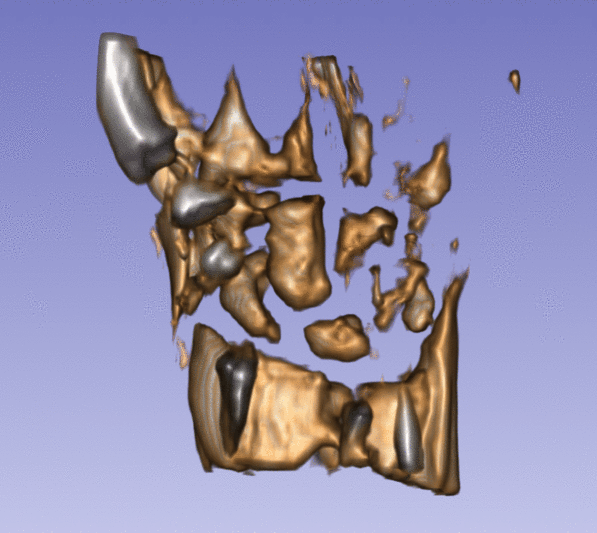
|
Segmentation of the wrist bones in the template space, which are propagated back to the individual observations via mappings computed during template estimation.
Continous change trajectories via spatiotemporal modeling. Color denotes speed and vectors denote direction of change.
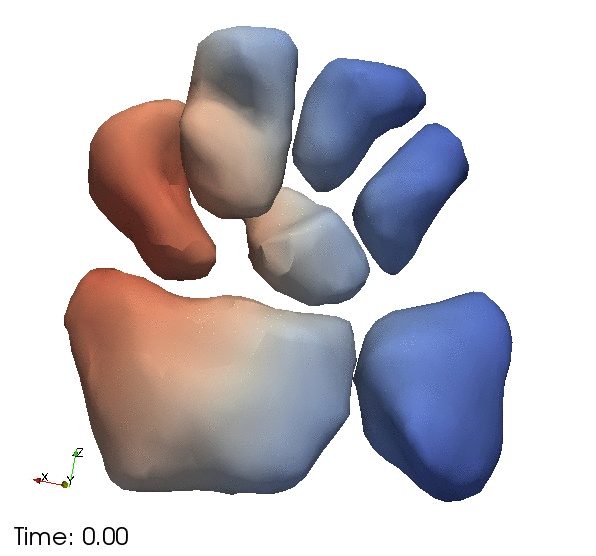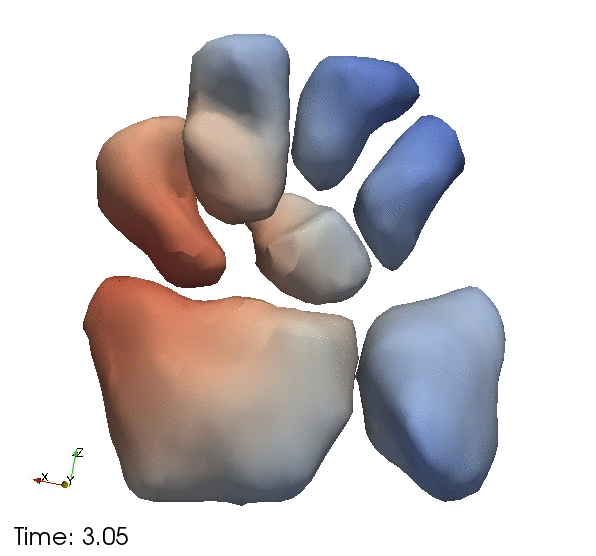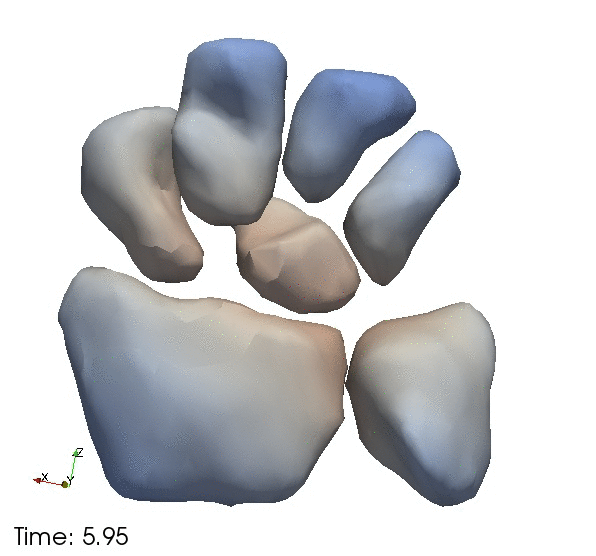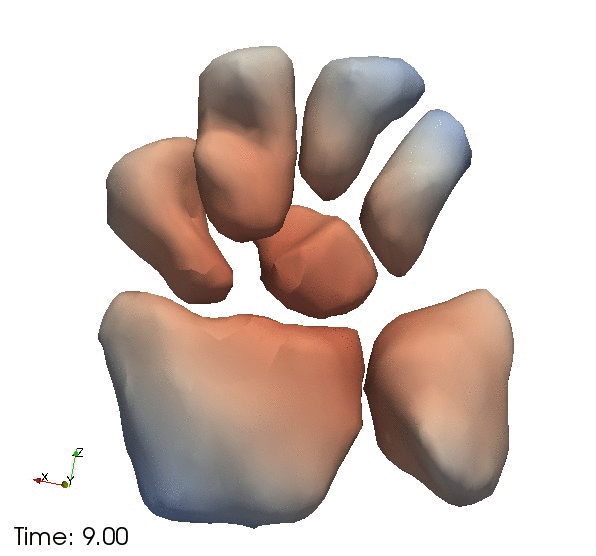
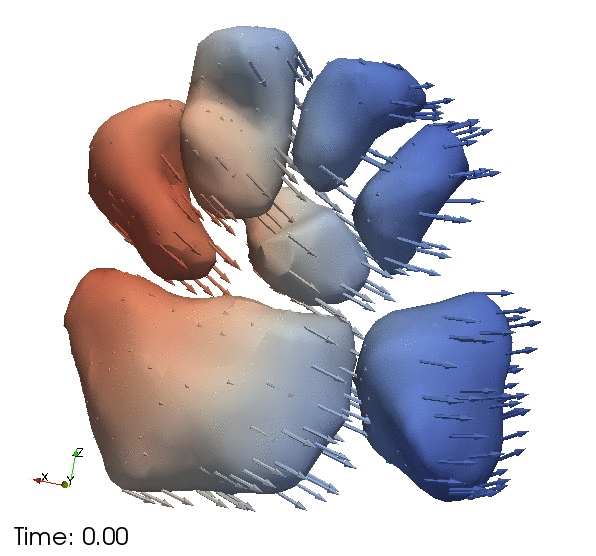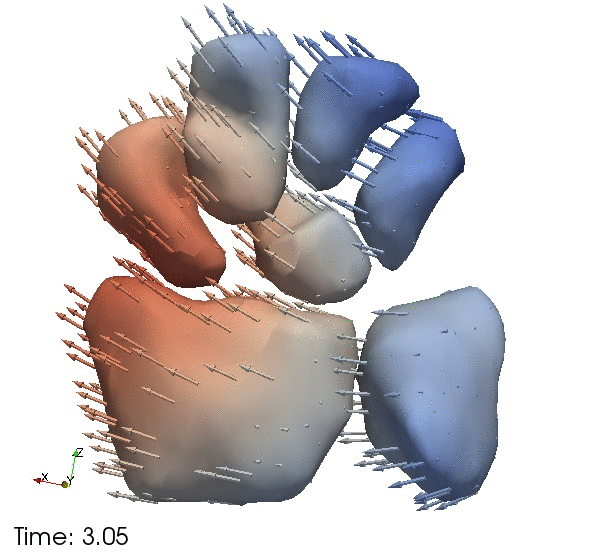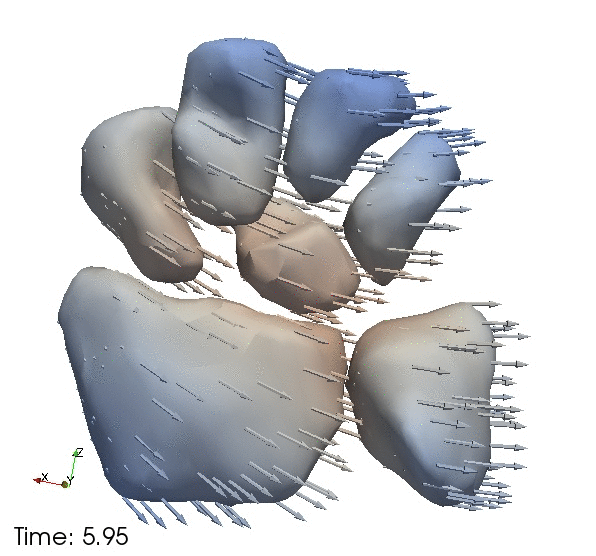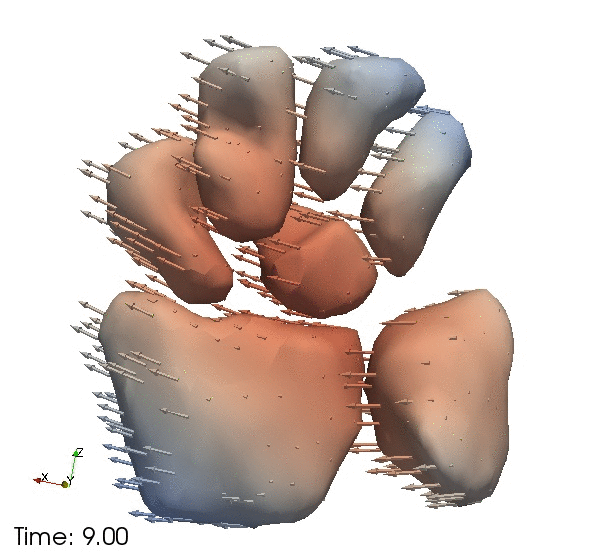
Background and References
[1] Fishbaugh, J., Durrleman, S., Prastawa, M., Gerig, G. Geodesic shape regression with multiple geometries and sparse parameters. Medical Image Analysis. Vol 39. pp. 1-17. (2017)
[2] Fishbaugh, J., Durrleman, S., Gerig, G. Estimation of smooth growth trajectories with controlled acceleration from time series shape data. Proc. of Medical Image Computing and Computer Assisted Intervention (MICCAI '11). Vol 6892, pp. 401-408. (2011)