Difference between revisions of "Projects/Diffusion/2007 Project Week Slicer 3 Whole Brain Seeding"
From NAMIC Wiki
| Line 1: | Line 1: | ||
{| | {| | ||
| − | |[[Image: | + | |[[Image:ProjectWeek-2007.png|thumb|320px|Return to [[2007_Programming/Project_Week_MIT|Project Week Main Page]] ]] |
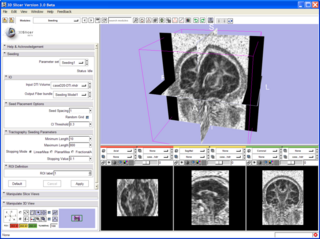
| − | |[[Image: | + | |[[Image:SeedingInterface.png|thumb|320px|Slicer 3 tractography display.]] |
|} | |} | ||
Revision as of 13:42, 29 June 2007
Home < Projects < Diffusion < 2007 Project Week Slicer 3 Whole Brain Seeding Return to Project Week Main Page |
Key Investigators
- BWH: Raul San Jose, Lauren O'Donnell, Alex Yarmarkovich
Objective
Deployment of whole brain seeding: data structure, fiber tracking and fast query mechanism to support editing operation and ROI-based fiber selection.
Approach, Plan
Our approach is to port the Streamline techniques used in Slicer 2 as a Command Line Module. We plan on looking at a optimal memory representation of a full brain tractography set for optimal querying and retrival of specific fibers according to spatial locations.
Progress
Software for the fiber tracking has been ported and interfaces has been updated to VTK 5. The command line module has been designed and fiber mrml bundles have been incorporated to the XML description (joint collaboration with Casey Goodlett).