Projects:AtlasBasedDTIFiberAnalyzerFramework
Back to UNC Algorithms
Atlas Based DTI Fiber Analysis Framework
This project aims to define an automatic framework for statistical comparison of fiber bundle diffusion properties between populations of diffusion weighted images.
Description
The general framework entails the following steps:
DWI and DTI quality control: DWI data suffers from inherent low SNR, overall long scanning time of multiple directional encoding with correspondingly large risk to encounter several kinds of artifacts. These artifacts can be too severe for a correct and stable estimation of the diffusion tensor. Thus, a quality control (QC) procedure is absolutely necessary for DTI studies. We developed a tool called DTIPrep which pipelines the QC steps with designated protocol use and report generation, and which generates DTI images and related scalar maps. More...
DTI preprocessing: skull-stripping
Skull-stripping is performed on DTI images and scalar maps. Several methods can be used in that regard:
- Direct Otsu Thresholding
- Masking using tissue label map from an intermediate atlas-based tissue segmentation, performed either on the idWI & B0 images or on the structural images (T1w & T2w).
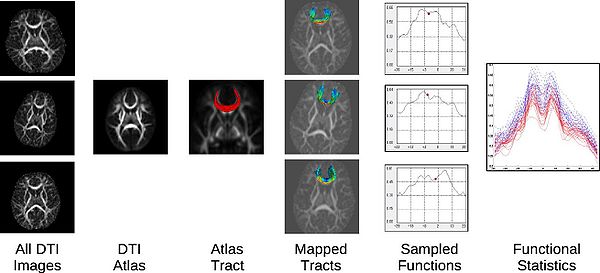
Unbiased DTI atlas building or atlas mapping: Unbiased diffeomorphic atlas building is used to compute a normalized coordinate system for populations of diffusion images. The diffeomorphic transformations between each subject and the atlas provide spatial normalization for the comparison of tract statistics.
- DTI atlas creation: A DTI atlas can be generated for a specific study by averaging all individual subjects. For longitudinal studies, a deformable longitudinal DTI atlas method can be used.
- DTI atlas mapping: An already existing DTI atlas can possibly be mapped to individual subjects. DTI-Reg can be used in that regard to perform DTI pairwise registration.
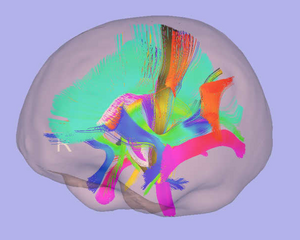
Tractography within 3D Slicer: Tractography is performed on the DTI atlas to generate tracts of interest. Several methods can be used in that regard:
- Single tensor-tractography Label seeding and ROI select
- Multi-tensor tractography with unscented kalman filter
Fiber cleanup/clustering:
Tracts generated on the DTI atlas often need to be cleaned up. FiberViewerLight enables several clustering methods: Length, Gravity, Hausdorff, and Mean methods but also a Normalized Cut algorithm.
DTIAtlasFiberAnalyzer:
Statistical analysis performed by statistician:
Merging statistics back to the original fiber bundle: MergeStatWithFiber - an application part of DTI Fiber Tracts Statistics package-, allows population statistical information to be merged back to the atlas fiber bundle.
3D visualization within 3D Slicer: Statically significan group differences can directly be displayed on a fiber bundle in 3D Slicer.
Publications
Key Investigators
- UNC Algorithms: Jean-Baptiste Berger, Benjamin Yvernault, Clement Vachet, Yundi Shi, Aditya Gupta, Martin Styner
- Utah Algorithms: Anuja Sharma, Sylvain Gouttard, Guido Gerig