Difference between revisions of "Projects:BayesianMRSegmentation"
| Line 17: | Line 17: | ||
We have validated the proposed technique by comparing our automated segmentation results with corresponding manual delineations in ultra-high resolution MRI scans (voxel size 0.38x0.38x0.80mm^3) of five individuals. For each of seven structures of interest (fimbria, CA1, CA2/3, CA4/DG, presubiculum, subiculum, and hippocampal fissure), we calculated the Dice overlap coefficient, defined as the volume of overlap between the automated and manual segmentation divided by their mean volume. We used a leave-one-out cross-validation strategy, in which we built an atlas mesh from the delineations in 4 subjects, and used this to segment the image of the remaining subject. We repeated this process for each of the 5 subjects, and compared the automated segmentation results with the corresponding manual delineations. | We have validated the proposed technique by comparing our automated segmentation results with corresponding manual delineations in ultra-high resolution MRI scans (voxel size 0.38x0.38x0.80mm^3) of five individuals. For each of seven structures of interest (fimbria, CA1, CA2/3, CA4/DG, presubiculum, subiculum, and hippocampal fissure), we calculated the Dice overlap coefficient, defined as the volume of overlap between the automated and manual segmentation divided by their mean volume. We used a leave-one-out cross-validation strategy, in which we built an atlas mesh from the delineations in 4 subjects, and used this to segment the image of the remaining subject. We repeated this process for each of the 5 subjects, and compared the automated segmentation results with the corresponding manual delineations. | ||
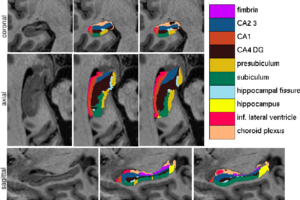
| − | Figure 1 compares the manual and automated segmentation results qualitatively on a set of cross-sectional slices. The upper half of figure 2 shows the average Dice overlap measure for each of the structures of interest, along with the minimum and maximum across the 5 subjects | + | Figure 1 compares the manual and automated segmentation results qualitatively on a set of cross-sectional slices. The upper half of figure 2 shows the average Dice overlap measure for each of the structures of interest, along with the minimum and maximum across the 5 subjects. The lower half of the figure shows, for each structure, the volume differences between the automated and manual segmentations relative to their mean volumes. An example of our mesh-based probabilistic atlas, derived from 4 manually labeled hippocampi, is shown in figure 3. |
| + | [[Image:MITHippocampalSubfieldSegmentationQualitative.png|thumb|center|300px|Fig 1. From left to right: ultra-high resolution MRI data, manual delineations, and corresponding automated segmentations.]] | ||
Revision as of 22:08, 16 May 2008
Home < Projects:BayesianMRSegmentationBack to NA-MIC Collaborations, MIT Algorithms
Model-Based Segmentation of Hippocampal Subfields in In Vivo MRI
Recent developments in MR data acquisition technology are starting to yield images that show anatomical features of the hippocampal formation at an unprecedented level of detail, providing the basis for hippocampal subfield measurement. Because of the role of the hippocampus in human memory and its implication in a variety of disorders and conditions, the ability to reliably and efficiently quantify its subfields through in vivo neuroimaging is of great interest to both basic neuroscience and clinical research. The aim of this project is to develop and validate a fully-automated method for segmenting the hippocampal subfields in ultra-high resolution MRI data.
Bayesian modeling and inference
We use a Bayesian modeling approach, in which we build an parametric computational model of how an MRI image around the hippocampal area is generated. The model incorporates a prior distribution that makes predictions about where neuroanatomical labels typically occur throughout the image, and is based on a generalization of probabilistic atlases that uses a deformable, compact tetrahedral mesh representation. The model also includes a likelihood distribution that predicts how a label image, where each voxel is assigned a unique neuroanatomical label, translates into an MRI image, where each voxel has an intensity.
Given an image to be segmented, we first estimate the parameters of our model that are most probable in light of the data. This parameter estimation involves finding the deformation that optimally warps the mesh-based probabilistic atlas onto the image under study, as well as finding the mean intensity and the intensity variance for each of the hippocampal subfields in the image. Once these parameters are estimated, the model is used to obtain the most probable image segmentation.
Results
We have validated the proposed technique by comparing our automated segmentation results with corresponding manual delineations in ultra-high resolution MRI scans (voxel size 0.38x0.38x0.80mm^3) of five individuals. For each of seven structures of interest (fimbria, CA1, CA2/3, CA4/DG, presubiculum, subiculum, and hippocampal fissure), we calculated the Dice overlap coefficient, defined as the volume of overlap between the automated and manual segmentation divided by their mean volume. We used a leave-one-out cross-validation strategy, in which we built an atlas mesh from the delineations in 4 subjects, and used this to segment the image of the remaining subject. We repeated this process for each of the 5 subjects, and compared the automated segmentation results with the corresponding manual delineations.
Figure 1 compares the manual and automated segmentation results qualitatively on a set of cross-sectional slices. The upper half of figure 2 shows the average Dice overlap measure for each of the structures of interest, along with the minimum and maximum across the 5 subjects. The lower half of the figure shows, for each structure, the volume differences between the automated and manual segmentations relative to their mean volumes. An example of our mesh-based probabilistic atlas, derived from 4 manually labeled hippocampi, is shown in figure 3.
Key Investigators
- MIT Algorithms: Koen Van Leemput, Polina Golland