Projects:ConnectivityAtlas
Back to NA-MIC Collaborations, MIT Algorithms,
Building an atlas of functional connectivity patterns
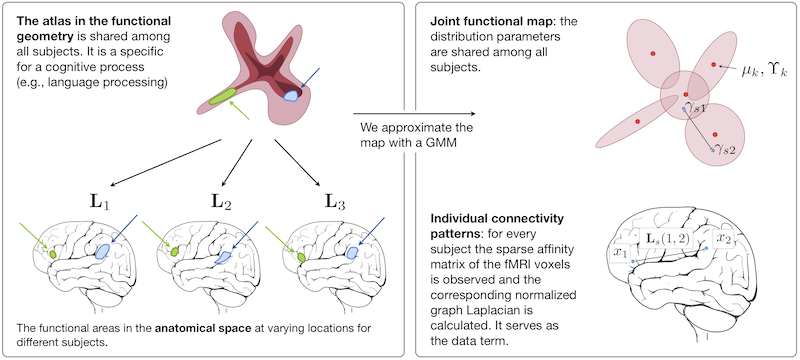
We construct an atlas that captures functional connectivity characteristics of a cognitive process from a population of individuals. The functional connectivity is encoded in a low-dimensional embedding space derived from a diffusion process on a graph that represents correlations of fMRI time courses. The atlas is represented by a common prior distribution for the embedded fMRI signals of all subjects. The atlas is not directly coupled to the anatomical space, and can represent functional networks that are variable in their spatial distribution. We derive an algorithm for fitting this generative model to the observed data in a population. Our results in a language fMRI study demonstrate that the method identifies coherent and functionally equivalent regions across subjects.
Description
We propose a different approach to characterize functional networks in a population of individuals. We do not assume a tight coupling between anatomical location and function, but view functional signals as the basis of a descriptive map that represents the global connectivity pattern during a specific cognitive process. We develop a representation of those networks based on manifold learning techniques and show how we can learn an \emph{atlas} from a population of subjects performing the same task. Our main assumption is that the connectivity pattern associated with a functional process is consistent across individuals. Accordingly, we construct a generative model (the atlas) for these connectivity patterns that describes the common structures within the population.
The clinical goal of this work is to provide additional evidence for localization of functional areas. A robust localization approach is important for neurosurgical planning if individual activations are weak or reorganization has happened due to pathologies such as tumor growth. Furthermore the method provides a basis for understanding the mechanisms underlying formation and reorganization in the cerebral system.
Experiments
Conclusion
Literature
[1] Georg Langs, Yanmei Tie, Laura Rigolo, Alexandra Golby, Polina Golland. Functional Geometry Alignment and Localization of Brain Areas. in Adv. in Neural Information Processing Systems NIPS 2010
[2] R.A. Heckemann, J.V. Hajnal, P. Aljabar, D. Rueckert, and A. Hammers. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage, 33(1):115–126, 2006.
[3] T. Rohlfing, R. Brandt, R. Menzel, and C.R. Maurer. Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. NeuroImage, 21(4):1428–1442, 2004.
[4] Freesurfer Wiki. http://surfer.nmr.mgh.harvard.edu.
Key Investigators
- MIT: Georg Langs, Andy Sweet, Danial Lashkari, Polina Golland
- Harvard: Yanmei Tie, Laura Rigolo, Alexandra Golby
Publications
NA-MIC Publications Database on Nonparametric Models for Supervised Image Segmentation