Projects:CorticalCorrespondenceWithParticleSystem
Back to NA-MIC Collaborations, UNC Algorithms, Utah Algorithms
Cortical Correspondence with Particle Systems
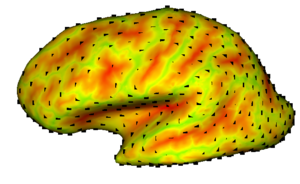
In this project, we want to compute cortical correspondence on populations, using various features such as cortical structure, DTI connectivity, vascular structure, and functional data (fMRI). This presents a challenge because of the highly convoluted surface of the cortex, as well as because of the different properties of the data features we want to incorporate together.
Description
We would like to use a particle based entropy minimizing system for the correspondence computation, in a population-based manner. This method best suits our needs since, parametrization based methods, such as MDL or SPHARM, require a spherical parametrization of the surface, which is hard to obtain for the highly convoluted cortex surface. Another advantage of using the particle based correspondence technique is that it does not require the surface to be of spherical topology; this means a lot less pre-processing for our method, since the brain cortex is not of spherical topology. Another strength of this method is that it would (eventually) enable correspondence computation on the subcortical structures and on the cortical surface using the same framework. We also would like to explore correspondence on the cerebellum, which traditionally is excluded from such studies (e.g. in FreeSurfer-based work).
The main disadvantage of using the particle based correspondence technique on the brain cortex is that it assumes the particles to be existing on local tangent planes, which presents a challenge for the cortex given the highly convoluted surface. We are hoping to overcome this difficulty by first ‘inflating’ the cortex surface. This way, we obtain a less convoluted, sphere-like surface, where the particles will be interacting. However, we need a 1-1 correspondence between the original cortex surface and the inflated surface, since the data to be used for correspondence, such as the the curvature, and the vascular data, lives on the original cortex surface. FreeSurfer offers a method that minimizes the distance distortion in the mapping, while also smoothing the surface. FreeSurfer also preprocesses the input surface to generate a spherical topology.
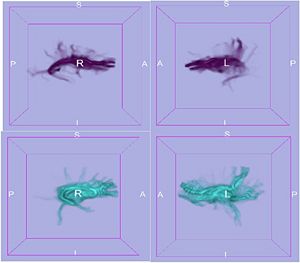
Once the framework for computing the correspondence given certain data features is established, the major challenge is to incorporate the various data forms that we would like to use together. We are planning on using structural data as well as connectivity (DTI). So far, we have tested our method using structural metrics, namely, sulcal depth (as computed by FreeSurfer), and have demonstrated improved correspondence quality compared to traditional, location-only correspondence (using the particle-based entropy framework), and we have shown that our results are at least comparable to FreeSurfer. This comes as no surprise as we had already shown in our previous studies that correspondence can be enhanced by using local curvature in addition of point locations for objects with complex geometry. Currently, we are at finalizing the use of connectivity information obtained with stochastic tractography methods developed as part of a separate NA-MIC project. In the image to the right, the cortical connectivity for the left and right caudate is visualized in 2 subjects.
Publications
- Oguz I, Cates J, Fletcher T, Whitaker R, Cool D, Aylward S, Styner M. Cortical correspondence using entropy-based particle systems and local features. Proceedings of the 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro 2008; 1637-1640.
- Cates J, Fletcher P, Styner M, Shenton M, Whitaker R. Shape modeling and analysis with entropy-based particle systems. Inf Process Med Imaging. 2007;20:333-45.
Key Investigators
- UNC Algorithms: Ipek Oguz, Martin Styner
- Utah Algorithms: Josh Cates, Tom Fletcher, Ross Whitaker